https://tiffanybtaylor.wordpress.com/

youtu.be/yv3Z8CZ3t_Q
#DNA #GeneticsForKids #ScienceBooks
advanced.onlinelibrary.wiley.com/doi/10.1002/...
We review nearly a decade of work asking:
how do gene regulatory networks rewire during evolution?
🧫🧵

advanced.onlinelibrary.wiley.com/doi/10.1002/...
We review nearly a decade of work asking:
how do gene regulatory networks rewire during evolution?
🧫🧵
Three SWBio DTP PhD project opportunities linked to the MultiDefence consortium are now open (1/4)
#PhD #DoctoralTraining #ResearchCarrers #LifeSciences #phagesky #AMR #microsky #PhDPositions
Three SWBio DTP PhD project opportunities linked to the MultiDefence consortium are now open (1/4)
#PhD #DoctoralTraining #ResearchCarrers #LifeSciences #phagesky #AMR #microsky #PhDPositions
✅ Depth over breadth
✅ Hands-on science
✅ Digital literacy
✅ Equity & progression.
My FREE Evolution Project brings these STEM priorities to life through engaging, hands-on learning.
www.bath.ac.uk/campaigns/di...
#STEM #CurriculumReview
✅ Depth over breadth
✅ Hands-on science
✅ Digital literacy
✅ Equity & progression.
My FREE Evolution Project brings these STEM priorities to life through engaging, hands-on learning.
www.bath.ac.uk/campaigns/di...
#STEM #CurriculumReview

doi.org/10.1098/rstb...

We are looking for a prospective PhD student to start in October 2026 who is excited about bacteria (Klebsiella pneumoniae), how they interact, and how they exchange DNA.
All details can be found on the funder website 👉 gw4biomed.ac.uk/developing-c...
We are looking for a prospective PhD student to start in October 2026 who is excited about bacteria (Klebsiella pneumoniae), how they interact, and how they exchange DNA.
All details can be found on the funder website 👉 gw4biomed.ac.uk/developing-c...
- Slow down
- Pay attention
- Visualise doing the whole protocol. What do you need to have labelled when? Where are you standing? What are you doing during the incubation? Etc.
- Start new methods with 2-3 small low-stakes experiments.
Good hands aren't some magic gift from the PCR gods, you have to develop them through directed repetitive practice, like any other skill
- Slow down
- Pay attention
- Visualise doing the whole protocol. What do you need to have labelled when? Where are you standing? What are you doing during the incubation? Etc.
- Start new methods with 2-3 small low-stakes experiments.
Read🧵for highlights and fun stats (1/13)

Read🧵for highlights and fun stats (1/13)
@ostratodd.bsky.social @philmuslab.bsky.social @aaronrashotte.bsky.social @taylorlabgroup.bsky.social Hao Chen, Matthew Cope-Arguello, Olufemi Isimikalu, Paul Scesa, K. Scott, S. Wakao, B. Woolston
jgi.doe.gov/user-science...

Could one classroom research experience be that catalyst?
Could a summer camp for teachers help them make students become scientists? 🧪🧫🧬🔬
YES. This is EvolvingSTEM. So proud of this team.
youtu.be/sv3tcwRs2PE?...

Could one classroom research experience be that catalyst?
Could a summer camp for teachers help them make students become scientists? 🧪🧫🧬🔬
YES. This is EvolvingSTEM. So proud of this team.
youtu.be/sv3tcwRs2PE?...
#phage #phagesky #microsky
Pseudomonads coordinate innate defense against viruses and bacteria with a single regulatory system:
www.cell.com/cell-host-mi...

#phage #phagesky #microsky
Pseudomonads coordinate innate defense against viruses and bacteria with a single regulatory system:
www.cell.com/cell-host-mi...
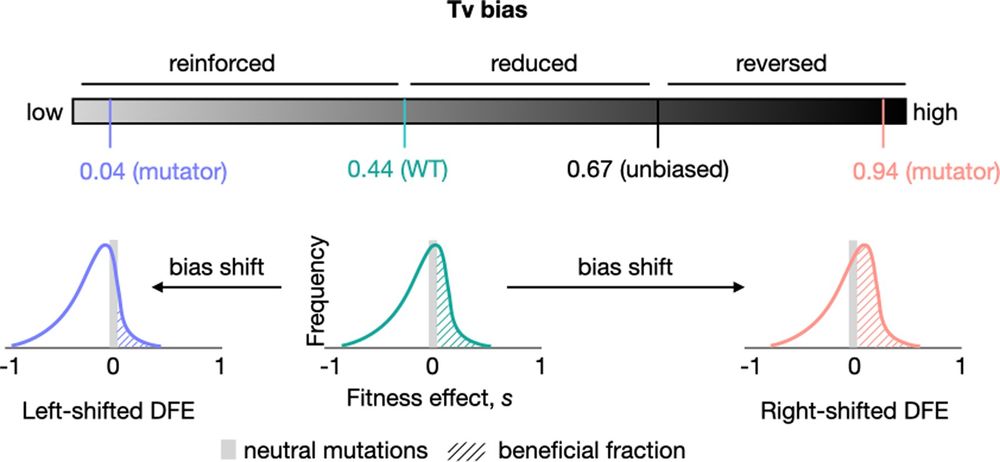
🔗 doi.org/10.1093/molbev/msaf183
#evobio #molbio

🔗 doi.org/10.1093/molbev/msaf183
#evobio #molbio
General across bacteria + potential in synbio.
📄 academic.oup.com/mbe/advance-ar…
🔗 James’s thread ⬇️
General across bacteria + potential in synbio.
📄 academic.oup.com/mbe/advance-ar…
🔗 James’s thread ⬇️
www.jobbnorge.no/en/available...

www.jobbnorge.no/en/available...




www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
wwwfbm.unil.ch/releve/appli...
www.tes.com/magazine/tea...
www.tes.com/magazine/tea...