Sudying histone modifications during programmed DNA elimination in Paramecium
PhD from @upcite.bsky.social @ijmonod.bsky.social

A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5

A collaborative effort spanning many years and several labs to uncover what the germline chromosomes of Paramecium really look like. 🔗 www.biorxiv.org/content/10.1...
1/5
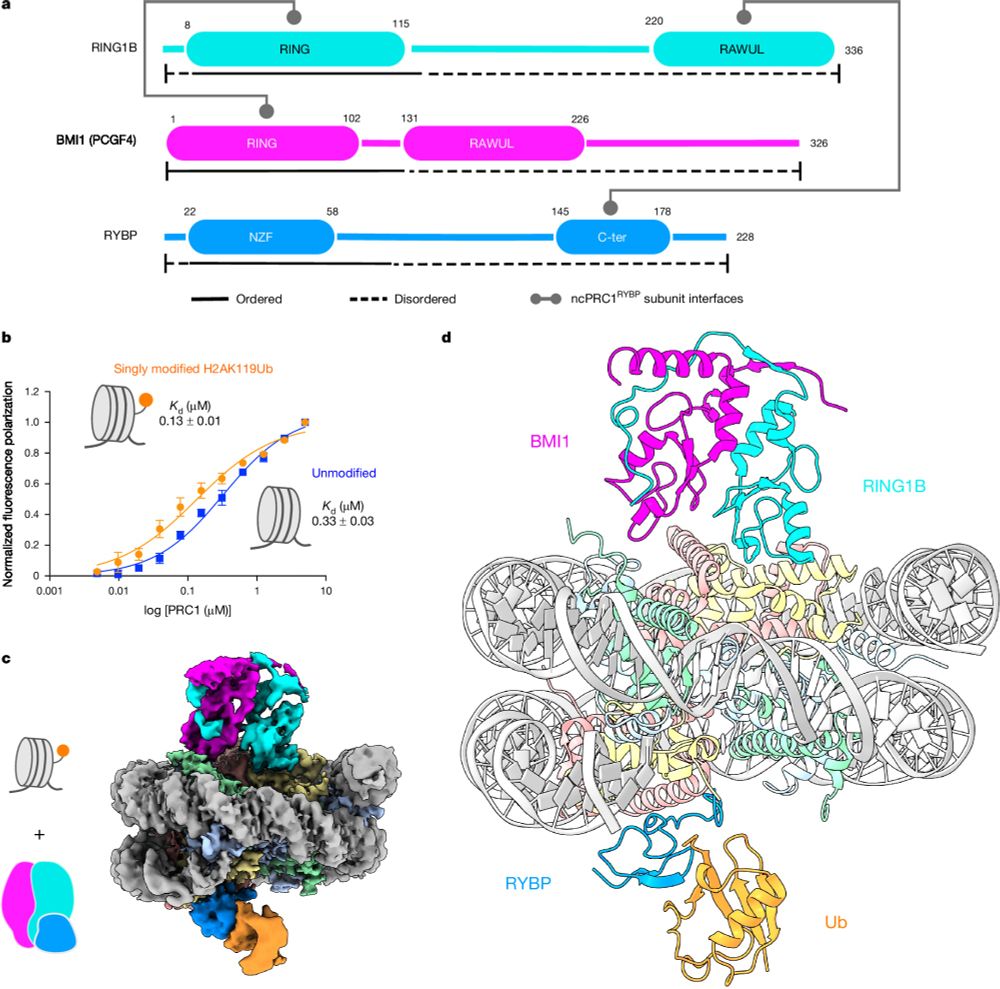
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...

This work is the fruit of a collaboration between the @sandraduharcourt.bsky.social & @betermieri2bc.bsky.social labs!
➡️ doi.org/10.1101/2025...
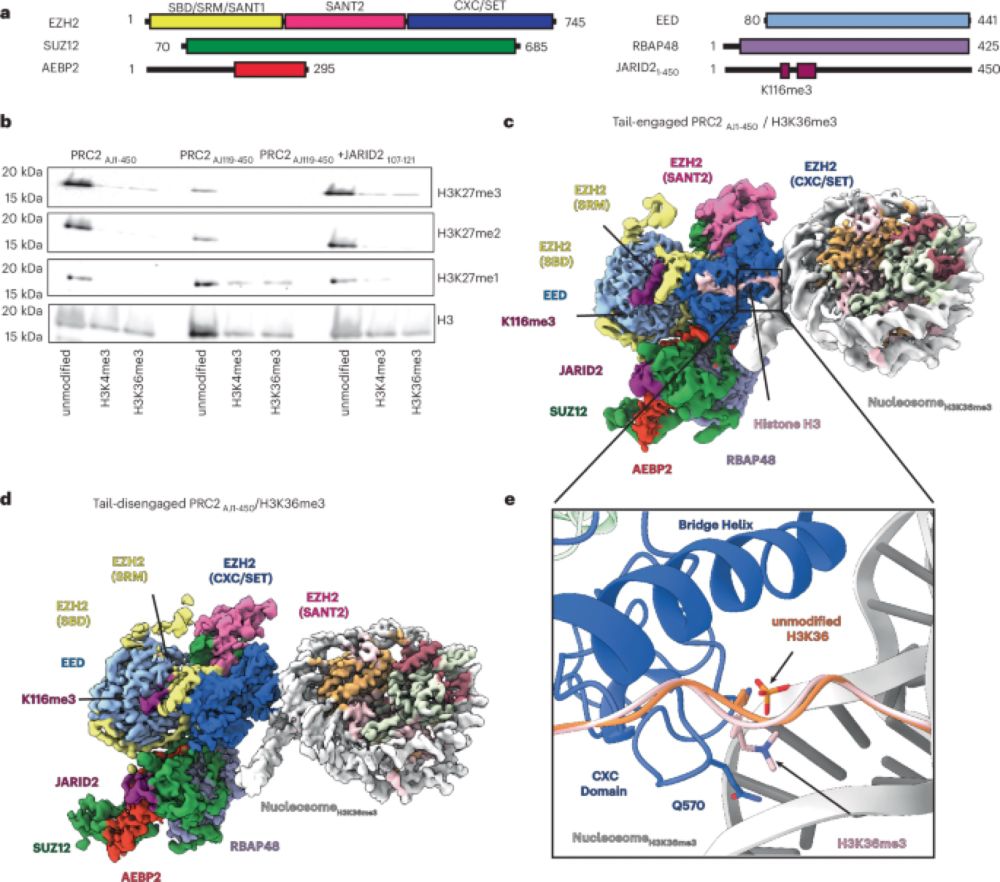
#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...

#RNAbiology #TEsky #smallRNAs #PRC2 #DNAelimination
1/5
academic.oup.com/nar/article-...
More informations 🔗 buff.ly/NhLWE6G
@cnrs-idf-villejuif.bsky.social @upcite.bsky.social

Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September

Interested in chromatin and its evolution? Good news! There's still time to join us in beautiful Catalonia (9-12 Dec) to discuss eukaryotic, bacterial, archaeal, and viral chromatin and how it all hangs together meetings.embo.org/event/24-evo...
Abstract deadline: 30 September
📅 Sept. 19th
📍 IJM
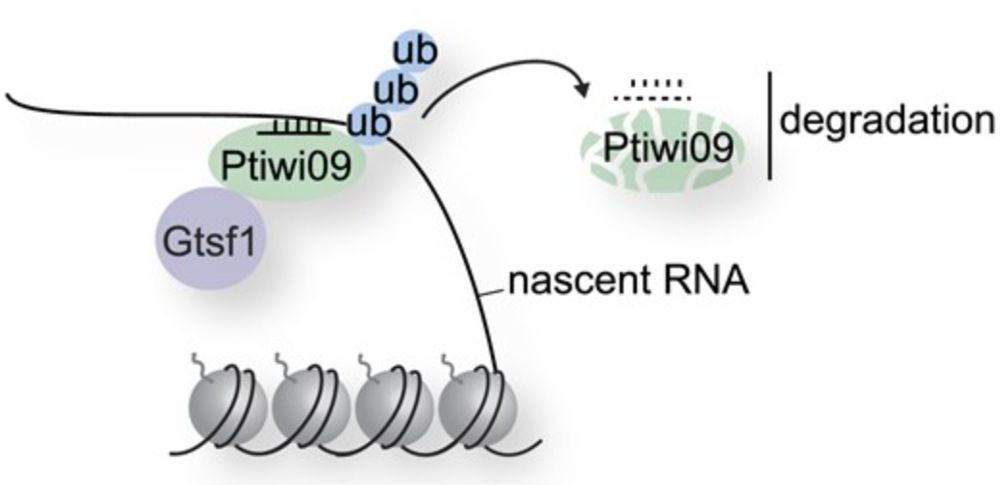
@thomasbalan.bsky.social ( @sandraduharcourt.bsky.social Lab ) will defend his PhD thesis "Characterization of proteins involved in the dynamics of histone modifications during programmed DNA elimination in Paramecium tetraurelia"
➡️ buff.ly/V8vzSsT

📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ

📅 September 18th
📍 Institut Jacques Monod
Invited by @sandraduharcourt.bsky.social’s lab, Peter Andersen ( @germline.bsky.social ) will give the IJM Seminar "Running to stand still – Recurrent innovation of animal germline genome regulation"
➡️ buff.ly/yaK2kTQ


www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM
www.biorxiv.org/content/10.1...
#Chromatin #Asgard #Archaea #cryoEM

But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
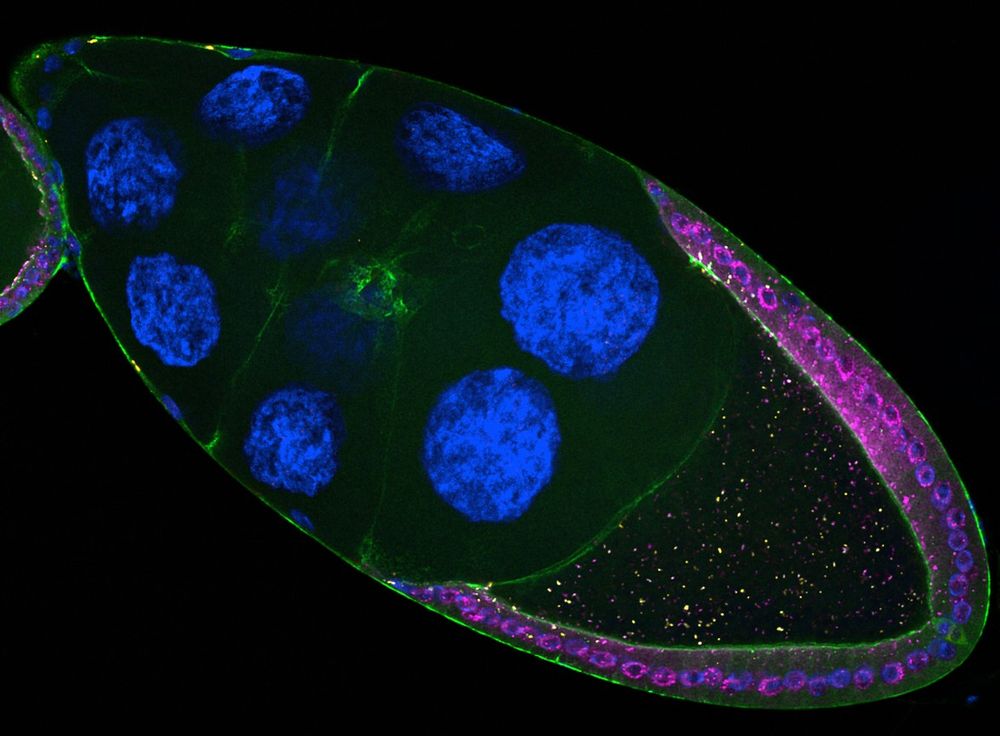
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
rdcu.be/d6Yly
rdcu.be/d6Yly

www.nature.com/articles/s41...
www.nature.com/articles/s41...

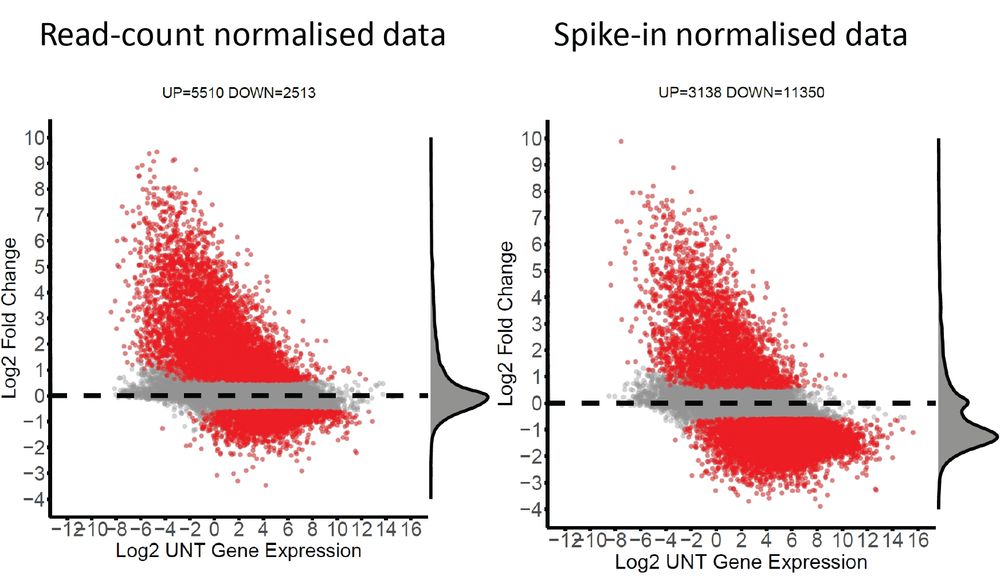
doi.org/10.1093/nar/...