www.imperial.ac.uk/news/271753/...

www.imperial.ac.uk/news/271753/...
Our kick-off meeting for the Klebsiella Seminar Series just wrapped up. Stay tuned for more!
@lauraamike.bsky.social @olayarendueles.bsky.social @caityholmes.bsky.social @tomstantonmicro.bsky.social @juanvalenciabacca.bsky.social WenWen Low & Jay V.
Our kick-off meeting for the Klebsiella Seminar Series just wrapped up. Stay tuned for more!
@lauraamike.bsky.social @olayarendueles.bsky.social @caityholmes.bsky.social @tomstantonmicro.bsky.social @juanvalenciabacca.bsky.social WenWen Low & Jay V.
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...
Topics include Kleb diversity, lineages, AMR, hypervirulence, how to use Kaptive & Kleborate for typing, and more!
klebnet.org/2025/11/18/k...

Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
Need a quick explainer on the new names? Check out my blog post: tinyurl.com/y8yb3rbb (+link to full review article)
#MicroSky @klebnet.bsky.social @tomstantonmicro.bsky.social
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
We aim to build a public metadata repository; systematic risk framework for global genomic surveillance; and genomic epi reviews for high-impact #Klebsiella clones.
Join us here:
klebnet.org/klebnet-gsp-...
#ABPHM25
www.rdocumentation.org/packages/jan...
www.rdocumentation.org/packages/jan...
📌The rapid detection of a neonatal unit outbreak of a wild-type Klebsiella variicola using decentralized Oxford Nanopore sequencing
doi.org/10.1186/s137...
@nanoporetech.com
🖥️🧬💻
#AcademicSky
#Microsky
🧪🧫🦠

📌The rapid detection of a neonatal unit outbreak of a wild-type Klebsiella variicola using decentralized Oxford Nanopore sequencing
doi.org/10.1186/s137...
@nanoporetech.com
🖥️🧬💻
#AcademicSky
#Microsky
🧪🧫🦠

But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
But this is just the beginning, we have lots of exciting things in store for the future of Kaptive to make in silico serotyping even better!
#kaptive #klebsiella #acinetobacter
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
Remember to cite us if you use Kaptive for your results, and watch out for "Untypeable"!
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
For the code-savvy, there's also a Python API allowing Kaptive to be used within your own programs 🧱
All the information you need is in the documentation, which we update regularly: kaptive.readthedocs.io/en/latest/
This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻

This means that if you don't have a fancy HPC, then don't worry! You can still analyse thousands of your own assemblies on your laptop in a reasonable time! 💻
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
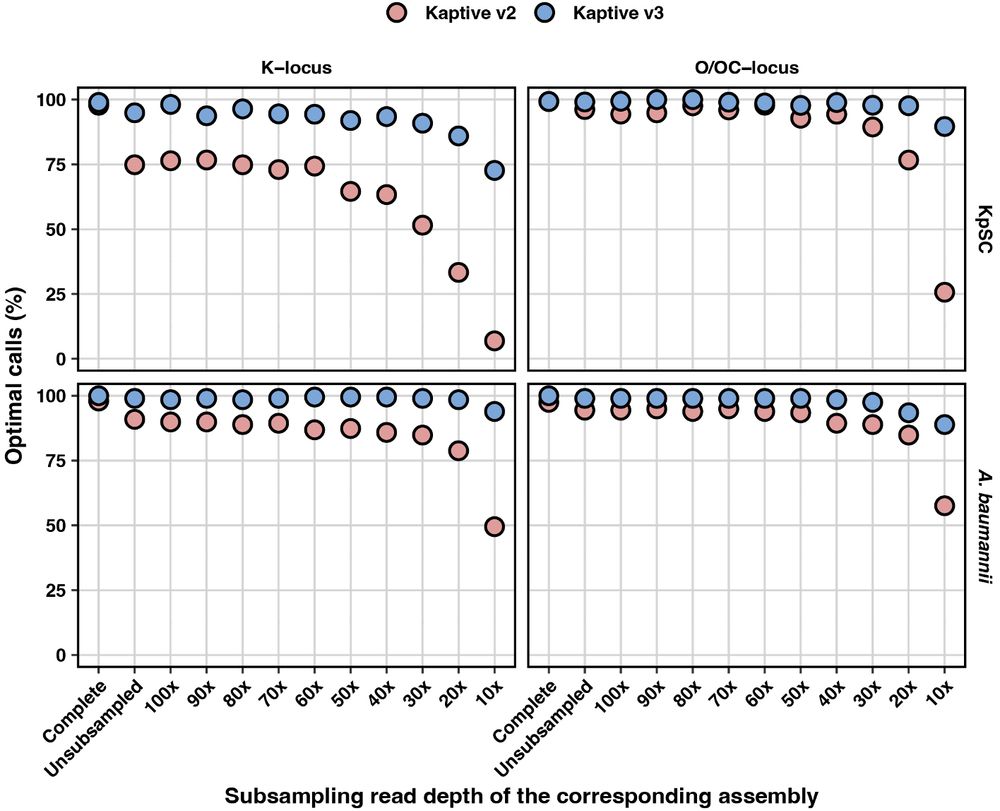
Kaptive 3 was much more sensitive than Kaptive 2, and maintained accuracy even when the assemblies were awful! 💩
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
We identified the K- and O(C)-loci in each and visually confirmed each to determine a ground truth Kaptive call 🔎
We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯

We also refactored (and simplified) the confidence score to be more sensitive for broken loci and missing genes, allowing more Kaptive data to be used when the assembly may not be complete 💯
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
Ever seen a stray KL107 in your data that didn't make sense?
Yeah, that's why...
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
The locus region is partially amplified ->
Low sequencing read coverage ->
region doesn't assemble well ->
Untypeable Kaptive call ->
Unusable data 🙅♀️
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
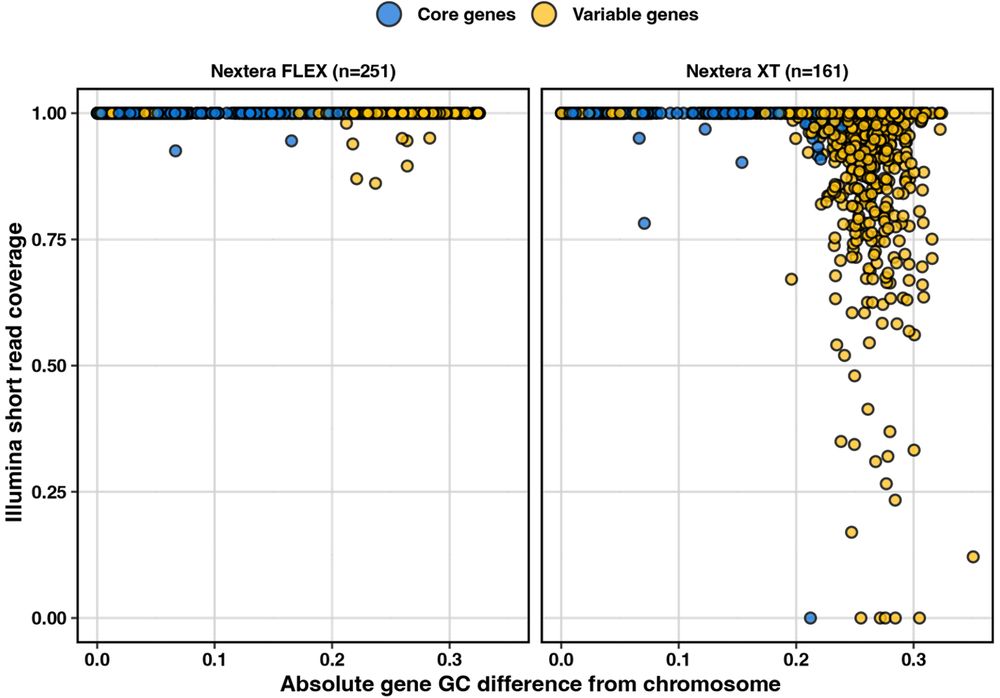
Turns out, these genes show decreased sequencing coverage when reads are prepped with Nextera XT, but not so much with Nextera Flex 🤯
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
Turns out, the genes missing were usually those important for antigenic diversity, in this case the glycosyltransferases that dictate the CPS 🍬 structure
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔
This meant that lots of useful seroepi data was unusable, so we started by finding out exactly why 🤔
Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...

Big thanks to coauthors @kelwyres.bsky.social, @katholt.bsky.social, @genomarit.bsky.social and Iren Löhr.
Here's what we did to improve in silico antigen typing 👇🧵
www.biorxiv.org/content/10.1...
Preprint now here: www.biorxiv.org/content/10.1...
Including a case study on nosocomial transmission of vancomycin resistant Enterococcus faecium
Preprint now here: www.biorxiv.org/content/10.1...
Including a case study on nosocomial transmission of vancomycin resistant Enterococcus faecium

