“the model knows something that cells don’t”
should be the guiding principle for making models.
“the model knows something that cells don’t”
should be the guiding principle for making models.
You might’ve found it challenging to tailor them to your specific use cases - not anymore!
Introducing MOFA-FLEX: a flexible, modular factor analysis framework designed for customizable modeling across diverse multi-omics data scenarios. 1/n

I generally do 2-5h in total and several times per day - across coding, science, career, life, health. It’s becoming quite integral. Hard to believe I would say this given what I thought before.
I generally do 2-5h in total and several times per day - across coding, science, career, life, health. It’s becoming quite integral. Hard to believe I would say this given what I thought before.

A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...

A rigorous mathematical foundation for Waddington's landscape to study cell fate decision making
Applied to ventral neural tube development
www.biorxiv.org/content/10.1...
@ribes
journals.plos.org/plosbiology/...

@ribes
journals.plos.org/plosbiology/...
We used genomic barcoding + scRNAseq in chick & human embryos to reveal a lineage architecture that reshapes how we understand neural tube development & cell fate decisions
🧵👇
www.biorxiv.org/content/10.1...

1/n
1/n
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
we present the first spatially resolved single cell atlas comparing radionecrotic changes (RN) and recurrent IDH-wildtype glioblastoma (GB) –– shedding light on a long-standing diagnostic challenge.
🧵 1/
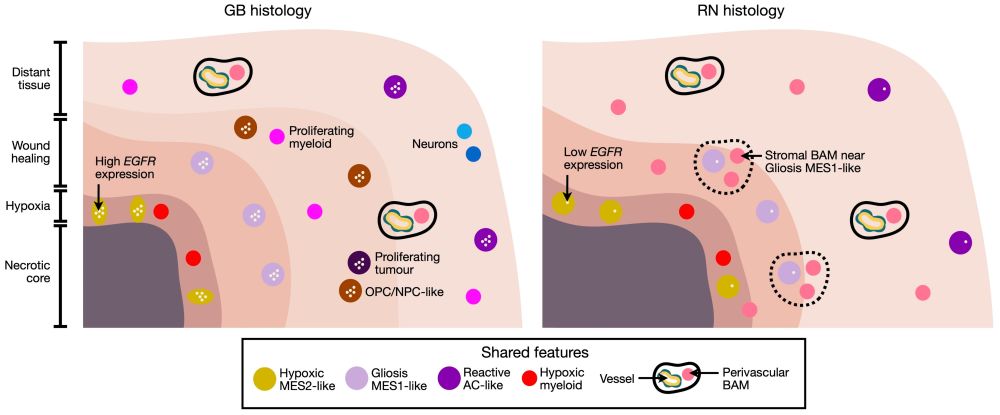
we present the first spatially resolved single cell atlas comparing radionecrotic changes (RN) and recurrent IDH-wildtype glioblastoma (GB) –– shedding light on a long-standing diagnostic challenge.
🧵 1/
Thinking about omnigenic model, “mechanistic bias” in medicine, personalised PRS.
Thinking about omnigenic model, “mechanistic bias” in medicine, personalised PRS.
www.cell.com/cell-metabol...

www.cell.com/cell-metabol...
www.biorxiv.org/content/10.1...
(1/n)

(6/n)


(6/n)
www.biorxiv.org/content/10.1...
(1/n)

Noise cancelling headphones are like your work email+slack - you may be able to work without them but it’s pretty hard.
Noise cancelling headphones are like your work email+slack - you may be able to work without them but it’s pretty hard.
If you can’t have windows open at night due to security concerns how are you supposed to ventilate?
I tried closing windows and even with the fan on max CO2 levels never got below 850ppm (bad for sleep quality).

If you can’t have windows open at night due to security concerns how are you supposed to ventilate?
I tried closing windows and even with the fan on max CO2 levels never got below 850ppm (bad for sleep quality).

To find out we built a #scRNAseq developmental atlas of the Drosophila nerve cord and linked it to the #connectome 🪰🧠
#preprint thread ⬇️1/8
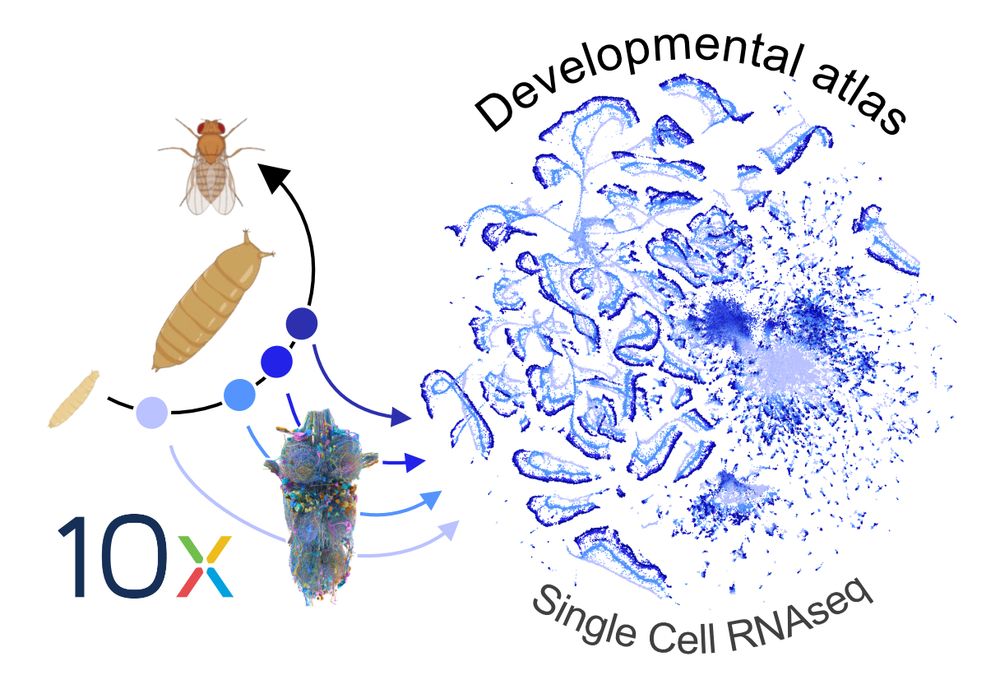
To find out we built a #scRNAseq developmental atlas of the Drosophila nerve cord and linked it to the #connectome 🪰🧠
#preprint thread ⬇️1/8

1. Wrong math
2. Wrong mapping of math to code (or vice versa)
3. Correct math and code but wrong interpretation given in text: code solves one task but paper presents it as another task
4. Poorly understood tasks
5. …
Linear mixed models and PCA can be described this way because as general methods they are wrong in less complex ways.
Specialised models can be wrong in quite misleading ways as well as useless.
1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
ESPOD builds on the collaborative relationship between EMBL-EBI and the @sangerinstitute.bsky.social, offering projects that combine experimental and computational approaches.
www.ebi.ac.uk/research/pos...
🧬🖥️🔬#postdocjob

1️⃣ Multicellular molecular characterisation of inflammatory bowel disease
6️⃣ Designing Efficient Single-Cell Perturbation Experiments with Lab-in-the-Loop AI Agents
📅 Application deadline is 30th September ❗
Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬

Find out how Tim’s group is exploring using large-scale single-cell and spatial data to trace cell lineages, understand cancer origins, and uncover how mutations drive disease.
www.ebi.ac.uk/about/news/p...
🖥️🧬

