London · briscoelab.org
For a summary & the digested highlights see thread🧵 from @giulia-boezio.bsky.social
www.biorxiv.org/content/10.1...
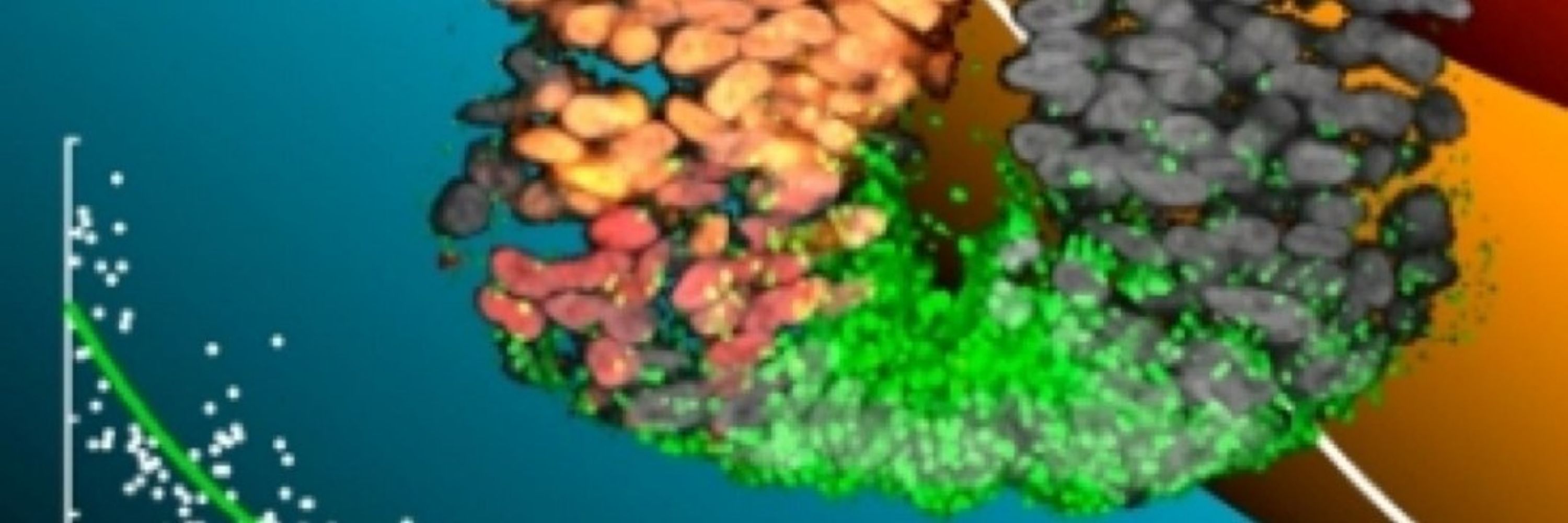
A computational framework for gene selection from single cell data that extracts biological signals in noisy data while avoiding artefacts from conventional dimensionality reduction
A thread
www.biorxiv.org/content/10.6...
A computational framework for gene selection from single cell data that extracts biological signals in noisy data while avoiding artefacts from conventional dimensionality reduction
A thread
www.biorxiv.org/content/10.6...
We'd love to hear from you: www.surveymonkey.com/r/GZCC9F9

We'd love to hear from you: www.surveymonkey.com/r/GZCC9F9
A computational framework for gene selection from single cell data that extracts biological signals in noisy data while avoiding artefacts from conventional dimensionality reduction
A thread
www.biorxiv.org/content/10.6...
A computational framework for gene selection from single cell data that extracts biological signals in noisy data while avoiding artefacts from conventional dimensionality reduction
A thread
www.biorxiv.org/content/10.6...
A few points and new information that bioscientists may find interesting...
A 🧵
A few points and new information that bioscientists may find interesting...
A 🧵
www.biologists.com/about-us/wor...

www.biologists.com/about-us/wor...
ESFS: A Noise-Resilient Framework for Feature Selection and Marker Gene Discovery in Single-Cell Transcriptomics | bioRxiv www.biorxiv.org/content/10.6...
ESFS: A Noise-Resilient Framework for Feature Selection and Marker Gene Discovery in Single-Cell Transcriptomics | bioRxiv www.biorxiv.org/content/10.6...
the St. Anna CCRI Symposium on Cell Fate in Cancer and Development tomorrow
ccri.at/st-anna-ccri...
the St. Anna CCRI Symposium on Cell Fate in Cancer and Development tomorrow
ccri.at/st-anna-ccri...
Recruiting soon: 4 postdocs + 3 PhDs
Press release in Italian — to decolonise scientific language 😄
magazine.unibo.it/it/articoli/...
Email me if interested in joining the lab

Recruiting soon: 4 postdocs + 3 PhDs
Press release in Italian — to decolonise scientific language 😄
magazine.unibo.it/it/articoli/...
Email me if interested in joining the lab
@maurazimmermann.bsky.social @maizel-lab.org @yusuke-mori.bsky.social @akankshi.bsky.social @nicolettapetridou.bsky.social

@maurazimmermann.bsky.social @maizel-lab.org @yusuke-mori.bsky.social @akankshi.bsky.social @nicolettapetridou.bsky.social
For those about to apply for their first independent academic position, the programme offers
- mentoring
- leadership training
- profile rising
- networking
journals.biologists.com/dev/pages/pi...
For those about to apply for their first independent academic position, the programme offers
- mentoring
- leadership training
- profile rising
- networking
journals.biologists.com/dev/pages/pi...
doi.org/10.1242/dev....

doi.org/10.1242/dev....
Editor-in-Chief @jamesbriscoe.bsky.social leads our team of Academic Editors in addressing the perception that Development is ‘too hard to publish in’ by discussing the journal's review process.
doi.org/10.1242/dev....

Editor-in-Chief @jamesbriscoe.bsky.social leads our team of Academic Editors in addressing the perception that Development is ‘too hard to publish in’ by discussing the journal's review process.
doi.org/10.1242/dev....


Thanks to the Indian SDB for hosting and to Sally Dunwoodie for the generous introduction.

Thanks to the Indian SDB for hosting and to Sally Dunwoodie for the generous introduction.
tinyurl.com/yc7u296j


tinyurl.com/yc7u296j
A chemo-optogenetic system for spatiotemporal control of gene expression
We use it to:
- Reconstruct SHH mediated neural tube patterning in vitro
- Measure extracellular half-life of Shh
www.cell.com/developmenta...

A chemo-optogenetic system for spatiotemporal control of gene expression
We use it to:
- Reconstruct SHH mediated neural tube patterning in vitro
- Measure extracellular half-life of Shh
www.cell.com/developmenta...
Speak to James about your next Development paper.

Catch up on where the 2022 PI fellows are now
Apply to become a 2026 PI fellow
journals.biologists.com/dev/article/...

Catch up on where the 2022 PI fellows are now
Apply to become a 2026 PI fellow
journals.biologists.com/dev/article/...
www.bbc.com/news/article...

www.bbc.com/news/article...
Applied to neural tube patterning, we show how morphogen signals reshape landscapes and drive fate decisions
www.pnas.org/doi/10.1073/...

Applied to neural tube patterning, we show how morphogen signals reshape landscapes and drive fate decisions
www.pnas.org/doi/10.1073/...
In this Review, Nikola Sekulovski, Amber E. Carleton, Chien-Wei Lin and Kenichiro Taniguchi (@taniguchi-lab.bsky.social) discuss recent advances in primate amnion development
doi.org/10.1242/dev....

In this Review, Nikola Sekulovski, Amber E. Carleton, Chien-Wei Lin and Kenichiro Taniguchi (@taniguchi-lab.bsky.social) discuss recent advances in primate amnion development
doi.org/10.1242/dev....
Mentoring
Profile raising
Leadership training
Network building
Spread the word...
www.biologists.com/grants/devel...

Mentoring
Profile raising
Leadership training
Network building
Spread the word...
www.biologists.com/grants/devel...
Mentoring
Profile raising
Leadership training
Network building
Spread the word...
www.biologists.com/grants/devel...

Mentoring
Profile raising
Leadership training
Network building
Spread the word...
www.biologists.com/grants/devel...

