Carolina Monzó
@carolinamonzo.bsky.social
140 followers
90 following
8 posts
PhD. 🏳️🌈
Immunology, aging and bioinformatics enthusiast. @MSCActions postdoc at @i2sysbio.bsky.social @csic.es
Posts
Media
Videos
Starter Packs
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
LongTREC
@longtrec.bsky.social
· Jul 13

📽️ From 🌞 Valencia: Meet Fabian Jetzinger in the LongTREC Series! | LongTREC
📽️ From 🌞 Valencia: Meet Fabian Jetzinger in the LongTREC Series!
Fabian is a Doctoral Candidate at BioBam Bioinformatics, working under the supervision of Stefan Götz, PhD. He is working to make the...
www.linkedin.com
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
I2SysBio
@i2sysbio.es
· May 9

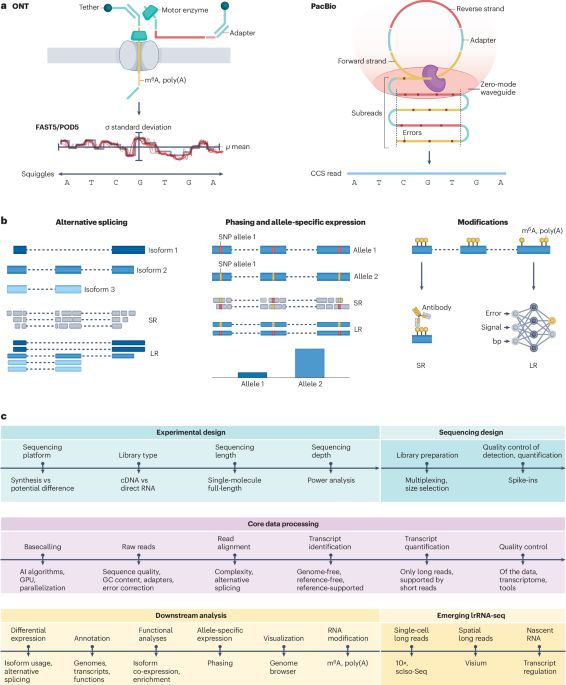
Transcriptomics in the era of long-read sequencing - Nature Reviews Genetics
Advances in long-read sequencing are driving the implementation of these technologies for transcriptome profiling. The authors provide a comprehensive guide to long-read RNA sequencing, including expe...
www.nature.com
Reposted by Carolina Monzó
I2SysBio
@i2sysbio.es
· May 9

I2SysBio explores a new gene sequencing technique to help decipher diseases such as cancer
<h1> </h1>
<h3><strong> This revision of the long-read transcriptomic sequencing technology allows a more precise analysis of the RNA molecules present in cells with essential information f...
i2sysbio.es
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó
Reposted by Carolina Monzó