DaiSyBio
@daisybio.de
42 followers
34 following
13 posts
Data Science in Systems Biology: We are a research group at the TUM School of Life Sciences. Cutting-edge expertise is united here in order to unlock the mechanisms of various systems in the human body.
Posts
Media
Videos
Starter Packs
Reposted by DaiSyBio
Reposted by DaiSyBio
Reposted by DaiSyBio
Reposted by DaiSyBio
Reposted by DaiSyBio
Tanja Laske
@mathbiotanja.bsky.social
· May 11
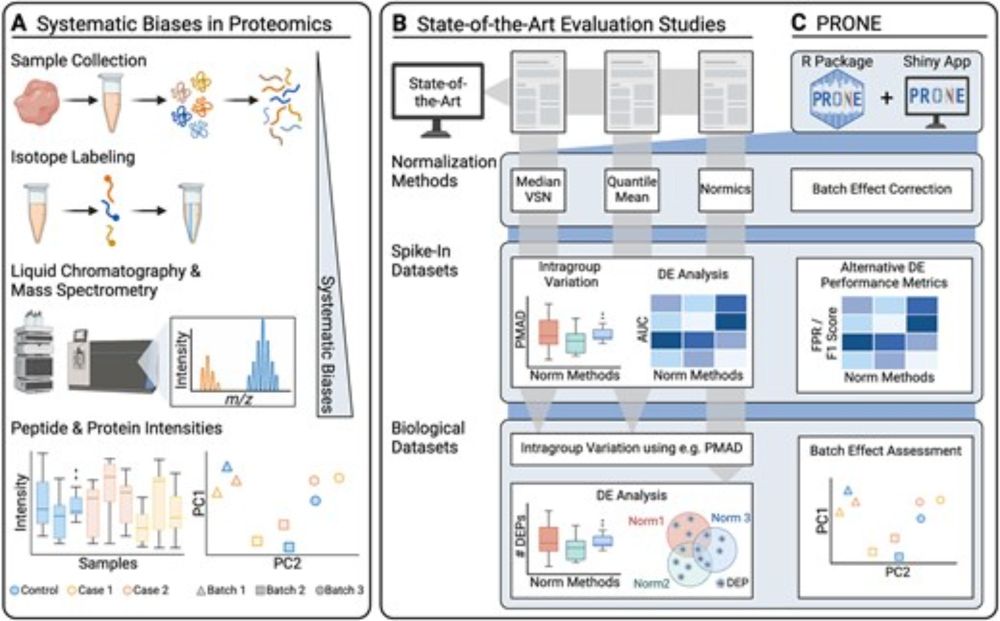
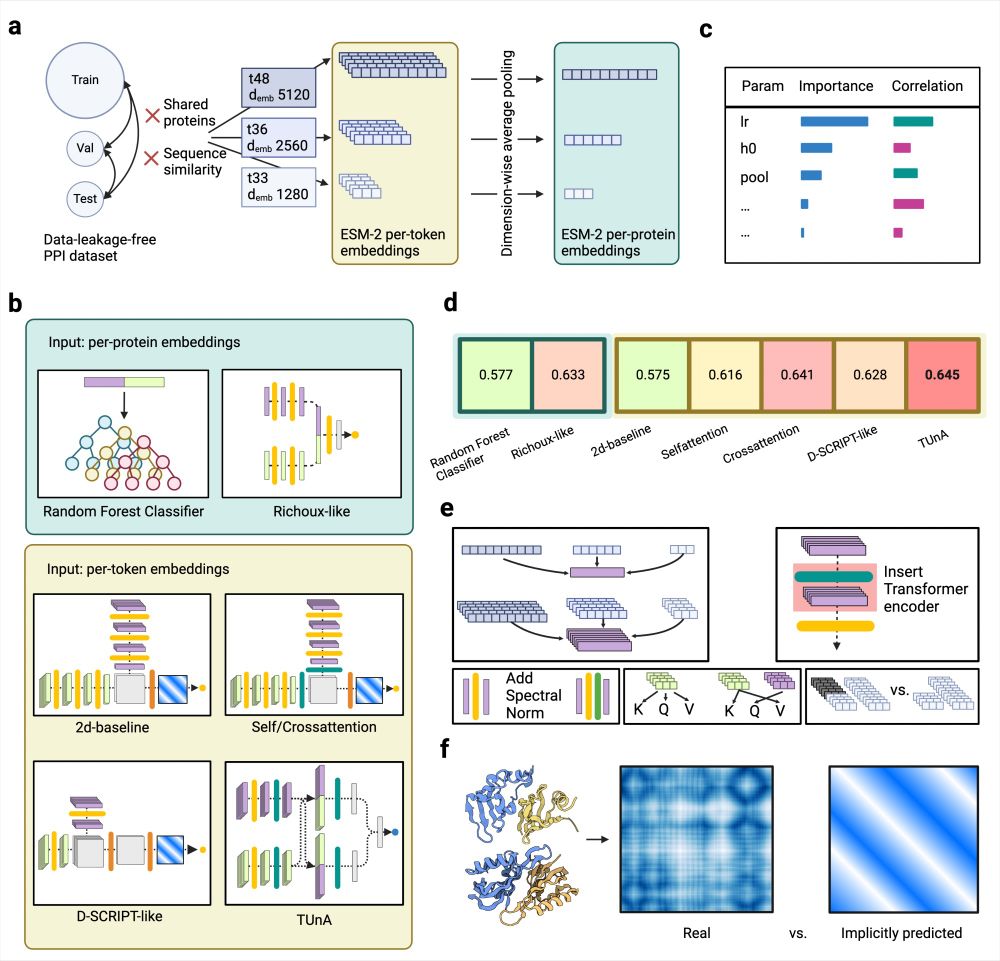
Systematic evaluation of normalization approaches in tandem mass tag and label-free protein quantification data using PRONE
Abstract. Despite the significant progress in accuracy and reliability in mass spectrometry technology, as well as the development of strategies based on i
doi.org
Reposted by DaiSyBio
Reposted by DaiSyBio
DaiSyBio
@daisybio.de
· Jan 30

Johannes Kersting Awarded an ASAPBio Poster Prize for RExPO Contribution
The ASAPBio poster competition is an initiative that champions open science by highlighting innovative research and the early sharing of results. This year’s competition emphasized transparency, colla...
blog.scienceopen.com
Reposted by DaiSyBio
Reposted by DaiSyBio