Alessandro Nicoli
@anicoli90.bsky.social
380 followers
400 following
14 posts
💻 Computational chemist who loves to understand how biomolecules interact at the molecular level
📍 Molecular Modeling Group at the Leibniz Institute at the Technical University of Munich (DE)
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Levental Lab
@leventallab.bsky.social
· May 7

Beyond Simple Models: The Consequences of Membrane Complexity in Living Systems - Copenhagen, Denmark 2025
Dear Colleagues,
We are excited to announce the Biophysical Society Thematic Meeting on Beyond Simple Models: The Consequences of Membrane Complexity in Living Systems. This meeting will be held in ...
www.biophysics.org
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli
Alessandro Nicoli
@anicoli90.bsky.social
· Mar 21
Reposted by Alessandro Nicoli
Bussi Lab
@bussilab.org
· Mar 18
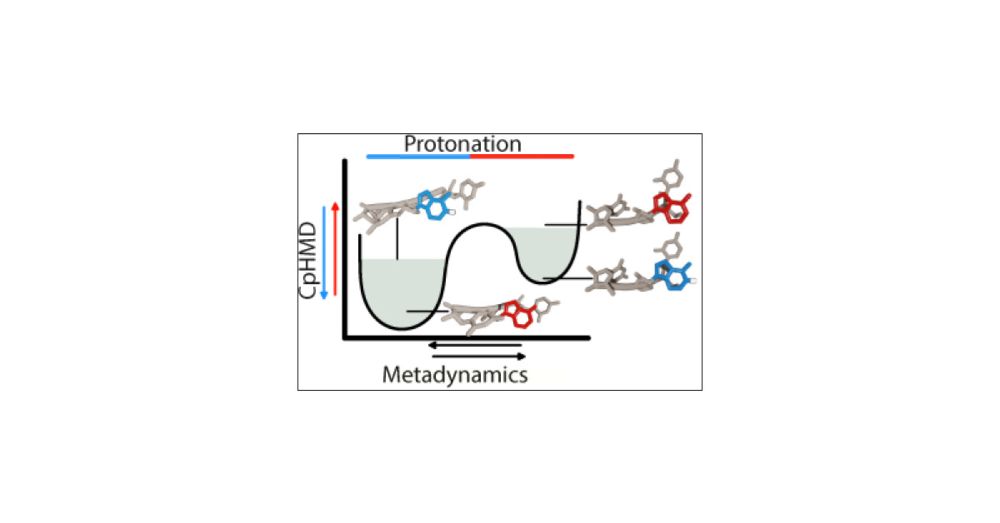
Characterizing RNA Oligomers Using Stochastic Titration Constant-pH Metadynamics Simulations
RNA molecules exhibit various biological functions intrinsically dependent on their diverse ecosystem of highly flexible structures. This flexibility arises from complex hydrogen-bonding networks defined by canonical and noncanonical base pairs that require protonation events to stabilize or perturb these interactions. Constant pH molecular dynamics (CpHMD) methods provide a reliable framework to explore the conformational and protonation spaces of dynamic structures and to perform robust calculations of pH-dependent properties, such as the pKa of titratable sites. Despite growing biological evidence concerning pH regulation of certain motifs and its role in biotechnological applications, pH-sensitive in silico methods have rarely been applied to nucleic acids. This work extends the stochastic titration CpHMD method to include RNA parameters from the standard χOL3 AMBER force field. We demonstrate its capability to capture titration events of nucleotides in single-stranded RNAs. We validate the method using trimers and pentamers with a single central titratable site while integrating a well-tempered metadynamics approach into the st-CpHMD methodology (CpH-MetaD) using PLUMED. This approach enhances the convergence of the conformational landscape and enables more efficient sampling of protonation-conformation coupling. Our pKa estimates are in agreement with experimental data, validating the method’s ability to reproduce electrostatic changes around a titratable nucleobase in single-stranded RNA. These findings provide molecular insight into intramolecular phenomena, such as nucleobase stacking and phosphate interactions, that dictate the experimentally observed pKa shifts between different strands. Overall, this work validates both the st-CpHMD method and the metadynamics integration as reliable tools for studying biologically relevant RNA systems.
doi.org
Reposted by Alessandro Nicoli
Reposted by Alessandro Nicoli