David E Sanin
@davidesanin.bsky.social
690 followers
520 following
26 posts
Assistant Professor at Johns Hopkins University and macrophage enthusiast! | he/him 🏳️🌈
Opinions are my own!
https://linktr.ee/davidesanin
Posts
Media
Videos
Starter Packs
Pinned
David E Sanin
@davidesanin.bsky.social
· Aug 22

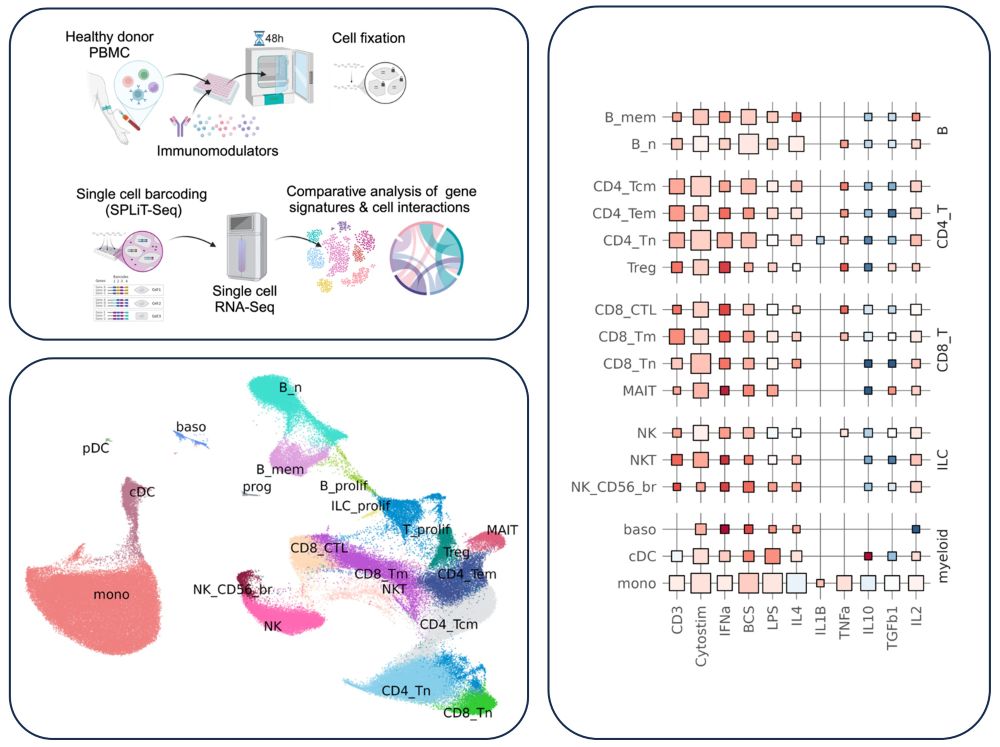
Missing data in single-cell transcriptomes reveals transcriptional shifts
Profiling thousands of single cell transcriptomes is routine, yet cell prioritization based on response to biological perturbations is challenging and confounded by clustering, normalization and dimen...
doi.org
David E Sanin
@davidesanin.bsky.social
· Aug 22
David E Sanin
@davidesanin.bsky.social
· Aug 22

Transcriptional shifts in scRNAseq revealed through Missingness
OAR (observed at random) score is a measure of transcriptional shifts among cells, allowing cell prioritization for downstream applications. For best results, test a group of similar cells where you e...
sanin-lab.github.io
David E Sanin
@davidesanin.bsky.social
· Aug 22

Diagnostic Test for Realized Missingness in Mixed-type Data - Sankhya B
A frequent concern in analyzing incomplete multivariate measurements in mixed categorical and quantitative scales is whether missing completely at random (MCAR) is an appropriate model. Realized MCAR ...
doi.org
David E Sanin
@davidesanin.bsky.social
· Aug 22

Missing data in single-cell transcriptomes reveals transcriptional shifts
Profiling thousands of single cell transcriptomes is routine, yet cell prioritization based on response to biological perturbations is challenging and confounded by clustering, normalization and dimen...
doi.org
David E Sanin
@davidesanin.bsky.social
· Jul 20

Regulation of inflammatory responses by pH-dependent transcriptional condensates
BRD4 functions as an intracellular pH sensor through a conserved histidine-rich intrinsically
disordered region. Inflammation-associated acidification triggers a pH-dependent switch
in transcriptional...
www.cell.com
Reposted by David E Sanin
Reposted by David E Sanin
Reposted by David E Sanin
David E Sanin
@davidesanin.bsky.social
· May 29

CoQ imbalance drives reverse electron transport to disrupt liver metabolism - Nature
Reverse electron transport is the mechanism behind excess mitochondrial reactive oxygen species in the livers of obese mice, which has implications for developing therapeutics for fatty liver dis...
www.nature.com
Reposted by David E Sanin
Denis Wirtz
@deniswirtz.bsky.social
· May 28
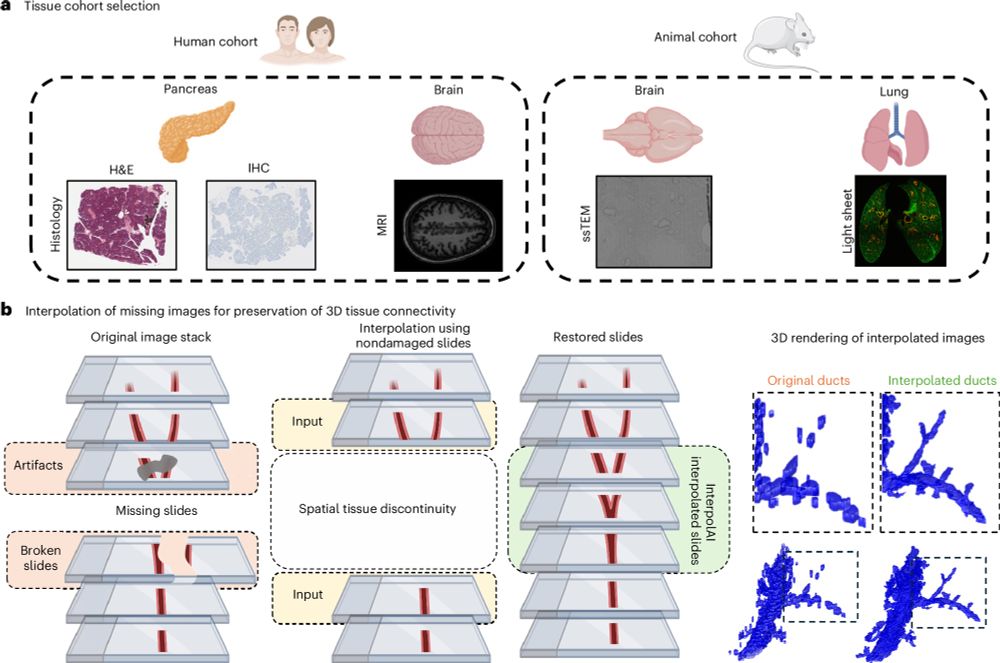
InterpolAI: deep learning-based optical flow interpolation and restoration of biomedical images for improved 3D tissue mapping
Nature Methods - InterpolAI leverages optimal flow-based artificial intelligence to produce synthetic images between pairs of images for diverse three-dimensional image types. InterpolAI is more...
www.nature.com
Reposted by David E Sanin
Reposted by David E Sanin
David E Sanin
@davidesanin.bsky.social
· Mar 16

Opinion | Musk Said No One Has Died Since Aid Was Cut. That Isn’t True. (Gift Article)
A journey through the front lines of global poverty shows that when the world’s richest men slash aid for the world’s poorest children, the result is sickness, starvation and death.
www.nytimes.com
Reposted by David E Sanin
Reposted by David E Sanin
Reposted by David E Sanin