Dominik Niopek
@dominikniopek.bsky.social
140 followers
48 following
9 posts
Science enthusiast. Synbio and Protein Enginnering Researcher. Professor @Heidelberg University. Enjoying drumming, guitar, hiking, biking, reading and, foremost, family & friends. All views are my own
Posts
Media
Videos
Starter Packs
Dominik Niopek
@dominikniopek.bsky.social
· Jun 13

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
www.biorxiv.org
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Dominik Niopek
@dominikniopek.bsky.social
· Jan 24
Reposted by Dominik Niopek
Rita Strack
@ritastrack.bsky.social
· Jan 23
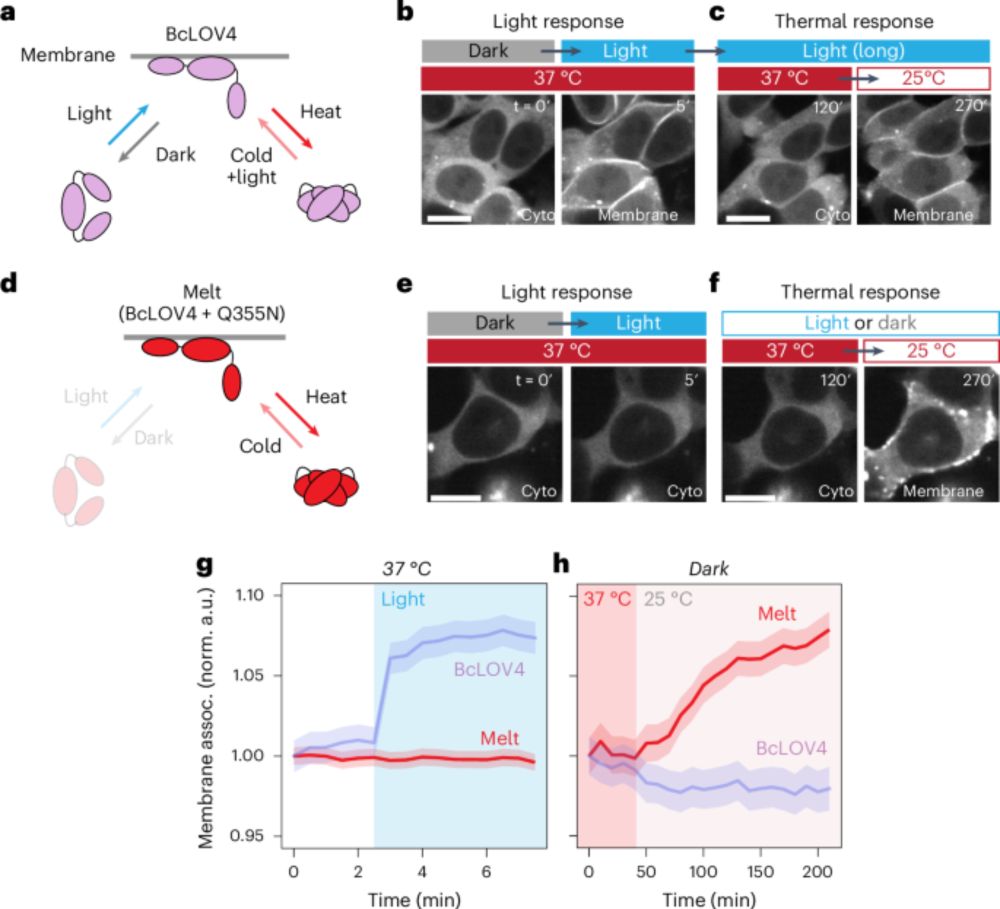
A temperature-inducible protein module for control of mammalian cell fate - Nature Methods
The Melt (Membrane localization using temperature) protein translocates to the plasma membrane upon temperature shift. Melt variants with a range of switching temperatures enable straightforward therm...
www.nature.com
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Kerstin Göpfrich
@kgoepfrich.bsky.social
· Dec 17

Full Professorship (W3) in Molecular Biology (f/m/d) with a Focus on Biological Engineering - Universität Heidelberg
Universität Heidelberg bietet Stelle als Full Professorship (W3) in Molecular Biology (f/m/d) with a Focus on Biological Engineering in Heidelberg - jetzt bewerben!
jobs.zeit.de
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Reposted by Dominik Niopek
Niopek Lab
@niopeklab.bsky.social
· Dec 5

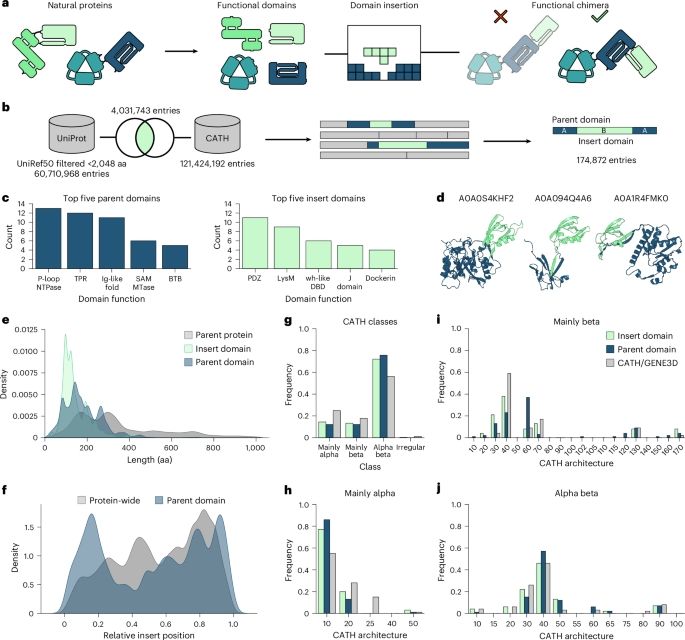
Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Domain insertion engineering is a powerful approach to juxtapose otherwise separate biological functions, resulting in proteins with new-to-nature activities. A prominent example are switchable protei...
www.biorxiv.org
Reposted by Dominik Niopek
Niopek Lab
@niopeklab.bsky.social
· Nov 26

A Versatile Anti-CRISPR Platform for Opto- and Chemogenetic Control of CRISPR-Cas9 and Cas12 across a Wide Range of Orthologs
CRISPR-Cas technologies have revolutionized life sciences by enabling programmable genome editing across diverse organisms. Achieving dynamic and precise control over CRISPR-Cas activity with exogenou...
www.biorxiv.org
Reposted by Dominik Niopek
Niopek Lab
@niopeklab.bsky.social
· Nov 26

A deep mutational scanning platform to characterize the fitness landscape of anti-CRISPR proteins
Abstract. Deep mutational scanning is a powerful method for exploring the mutational fitness landscape of proteins. Its adaptation to anti-CRISPR proteins,
doi.org