Jan Mathony
@jmathony.bsky.social
93 followers
110 following
22 posts
Scientist with a passion for Synbio | Protein engineering | Optogenetics | ML
www.niopeklab.de/mathony-lab/
Posts
Media
Videos
Starter Packs
Reposted by Jan Mathony
Jan Mathony
@jmathony.bsky.social
· Aug 22
Jan Mathony
@jmathony.bsky.social
· Aug 5
Reposted by Jan Mathony
Jan Mathony
@jmathony.bsky.social
· Aug 4

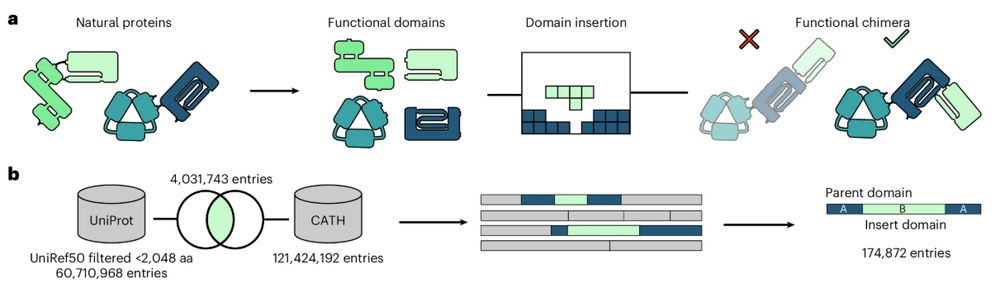
Rational engineering of allosteric protein switches by in silico prediction of domain insertion sites
Nature Methods - ProDomino is a machine leaning-based method, trained on a semisynthetic domain insertion dataset, to guide the engineering of protein domain recombination.
rdcu.be
Reposted by Jan Mathony
Reposted by Jan Mathony
Jan Mathony
@jmathony.bsky.social
· Jun 20
Reposted by Jan Mathony
Michael Jendrusch
@mjendrusch.bsky.social
· Jun 19
Reposted by Jan Mathony
Niopek Lab
@niopeklab.bsky.social
· Jun 13

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
doi.org
Jan Mathony
@jmathony.bsky.social
· Jun 13
Niopek Lab
@niopeklab.bsky.social
· Jun 13

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
doi.org
Jan Mathony
@jmathony.bsky.social
· Jun 13
Dominik Niopek
@dominikniopek.bsky.social
· Jun 13

Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
www.biorxiv.org
Jan Mathony
@jmathony.bsky.social
· May 3

Modular Engineering of Thermo-Responsive Allosteric Proteins
Thermogenetics enables non-invasive spatiotemporal control over protein activity in living cells and tissues, yet its applications have largely been restricted to transcriptional regulation and membra...
www.biorxiv.org
Jan Mathony
@jmathony.bsky.social
· May 3
Jan Mathony
@jmathony.bsky.social
· May 3

Precision control of cellular functions with a temperature-sensitive protein - Nature Methods
Temperature-sensitive proteins would enable the remote control of cellular functions deep within tissues, although few such proteins have been characterized. Melt is a protein that reversibly clusters...
www.nature.com
Jan Mathony
@jmathony.bsky.social
· May 3
Jan Mathony
@jmathony.bsky.social
· May 3
Jan Mathony
@jmathony.bsky.social
· Apr 22
Reposted by Jan Mathony
Kerstin Göpfrich
@kgoepfrich.bsky.social
· Apr 20

PyFuRNAce: An integrated design engine for RNA origami
To realize the full potential of RNA nanotechnology and RNA origami, user-friendly design tools are needed. Here, we present pyFuRNAce, an open-source, Python-based software package with a graphical u...
www.biorxiv.org
Reposted by Jan Mathony
Reposted by Jan Mathony
Reposted by Jan Mathony
Kaessmann Lab
@kaessmannlab.bsky.social
· Feb 13

Developmental origins and evolution of pallial cell types and structures in birds
Innovations in the pallium likely facilitated the evolution of advanced cognitive abilities in birds. We therefore scrutinized its cellular composition and evolution using cell type atlases from chick...
www.science.org