Dr Gareth Hawkes
@drghawkes.bsky.social
350 followers
380 following
24 posts
MRC Career Development Fellow, Lecturer in Health Data Science, University of Exeter. Academic in genetics, rare variants and complex traits
Posts
Media
Videos
Starter Packs
Reposted by Dr Gareth Hawkes
Tim Frayling
@timfrayling.bsky.social
· May 27
Dr Gareth Hawkes
@drghawkes.bsky.social
· May 27
Reposted by Dr Gareth Hawkes
Reposted by Dr Gareth Hawkes
Jeff Spence
@jeffspence.github.io
· Mar 28

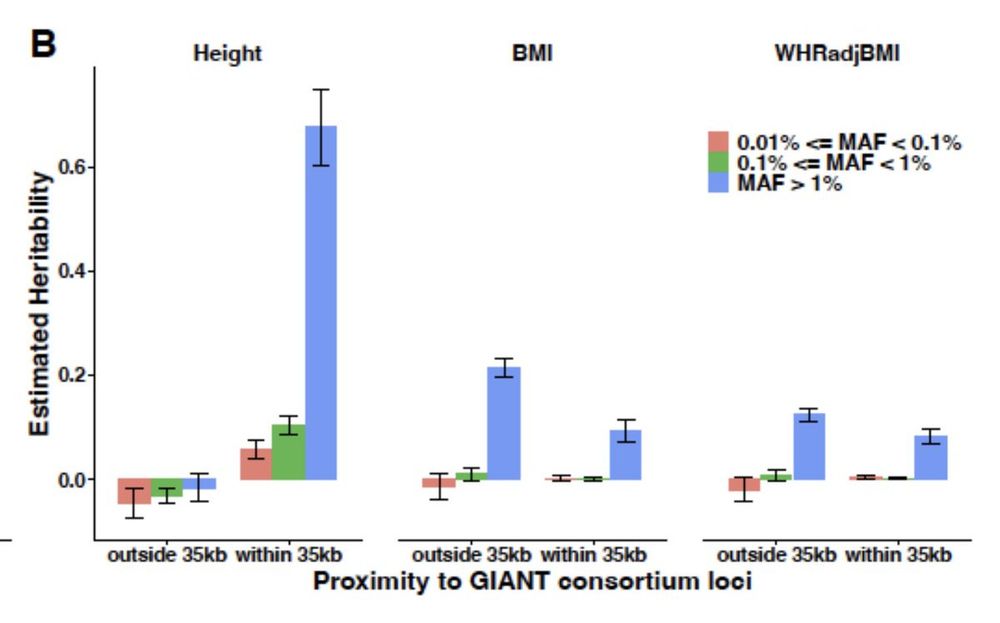
Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations
Genetic association studies have mostly focussed on common variants from genotyping arrays or rare protein-coding variants from exome sequencing. Here, we used whole-genome sequence (WGS) data in 672,...
www.biorxiv.org
Dr Gareth Hawkes
@drghawkes.bsky.social
· Mar 24
Reposted by Dr Gareth Hawkes
Reposted by Dr Gareth Hawkes
Reposted by Dr Gareth Hawkes
Mike Inouye
@mikeinouye.bsky.social
· Feb 25

Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations
Genetic association studies have mostly focussed on common variants from genotyping arrays or rare protein-coding variants from exome sequencing. Here, we used whole-genome sequence (WGS) data in 672,...
www.biorxiv.org
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 26

Whole-genome sequencing analysis of anthropometric traits in 672,976 individuals reveals convergence between rare and common genetic associations
Genetic association studies have mostly focussed on common variants from genotyping arrays or rare protein-coding variants from exome sequencing. Here, we used whole-genome sequence (WGS) data in 672,...
www.biorxiv.org
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 24
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 24
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 24
Dr Gareth Hawkes
@drghawkes.bsky.social
· Feb 24