Eimear Lagan
@eimearlagan.bsky.social
54 followers
96 following
13 posts
Post-doc @ IGC, University of Edinburgh
Epigenetics | Paediatric cancer | Functional genomics
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Eimear Lagan
Eimear Lagan
@eimearlagan.bsky.social
· Jun 7
Reposted by Eimear Lagan
Jongmin Kim
@jongminkmg.bsky.social
· Jun 6
Eimear Lagan
@eimearlagan.bsky.social
· May 21
Reposted by Eimear Lagan
Molecular Cell
@cp-molcell.bsky.social
· May 21
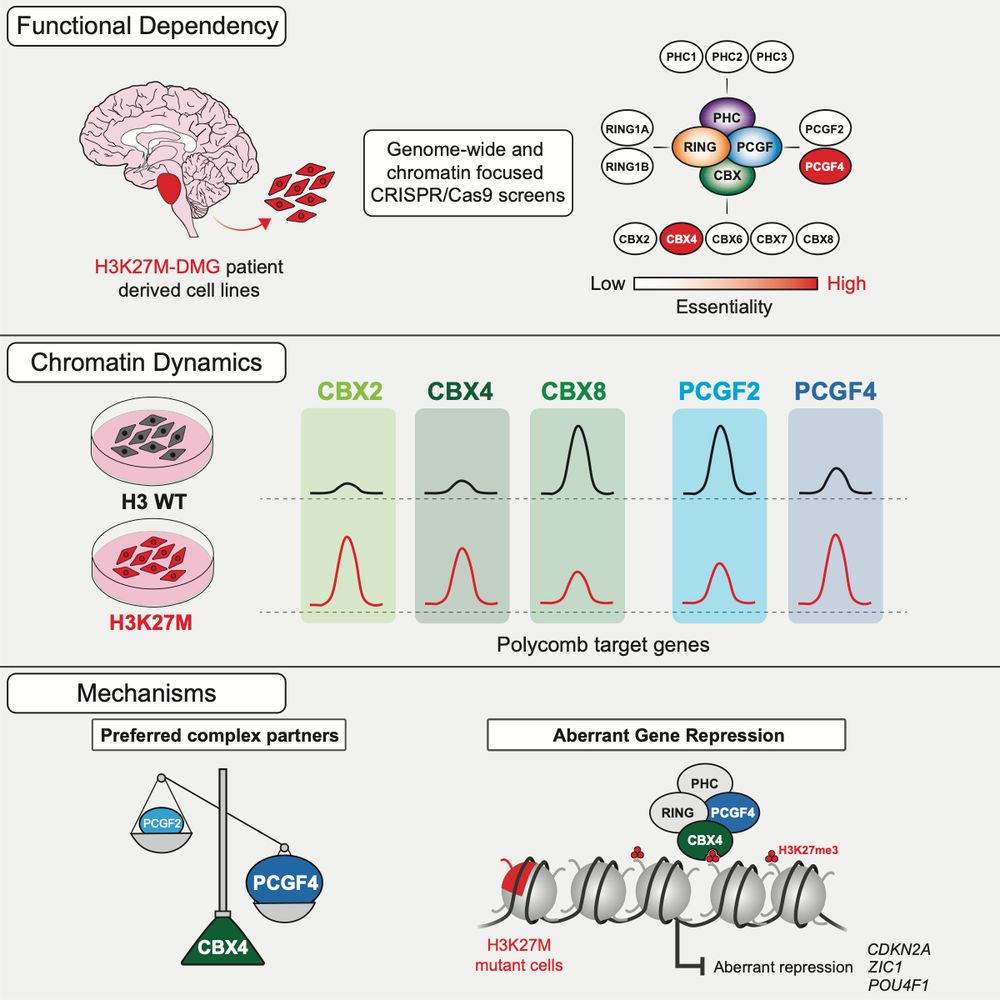
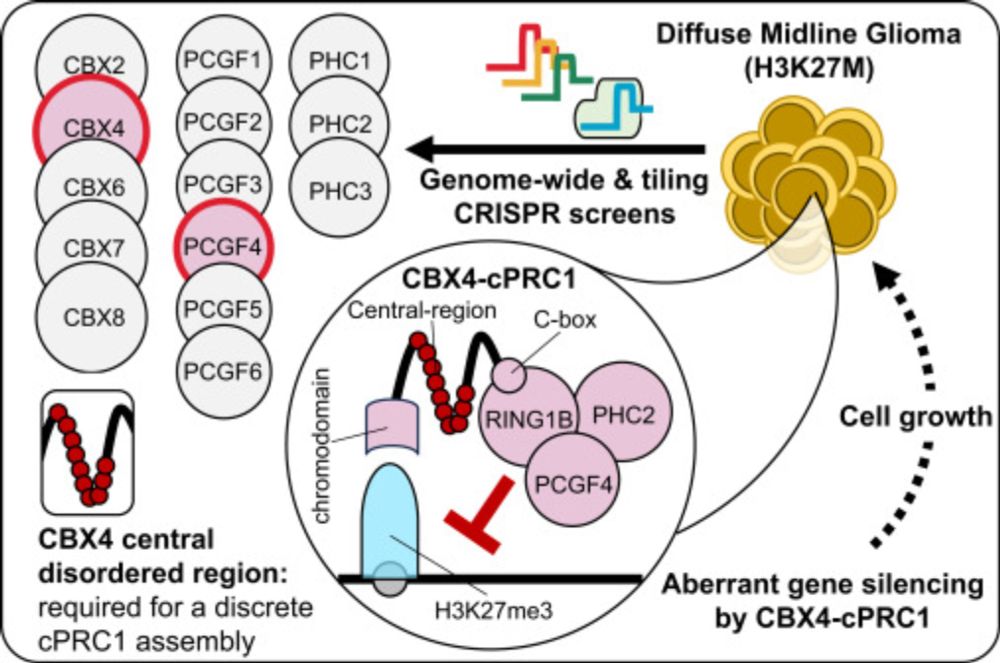
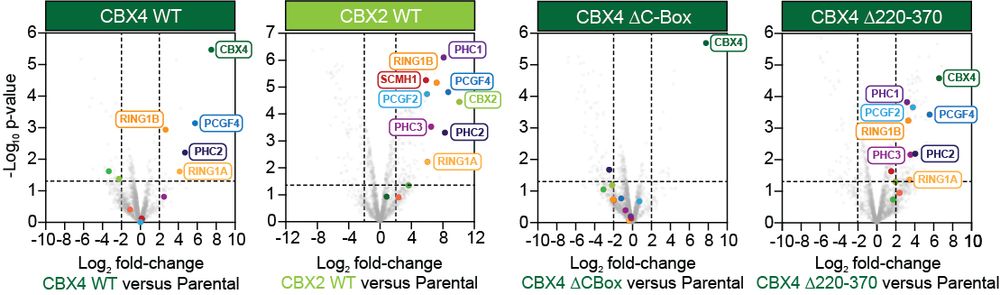
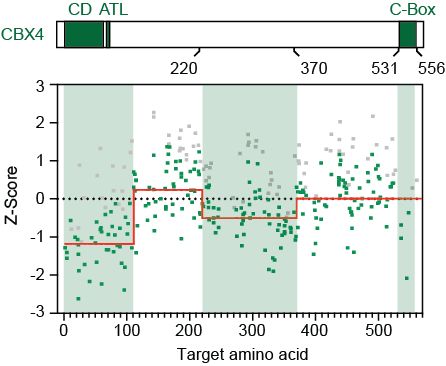
A specific form of cPRC1 containing CBX4 is co-opted to mediate oncogenic gene repression in diffuse midline glioma
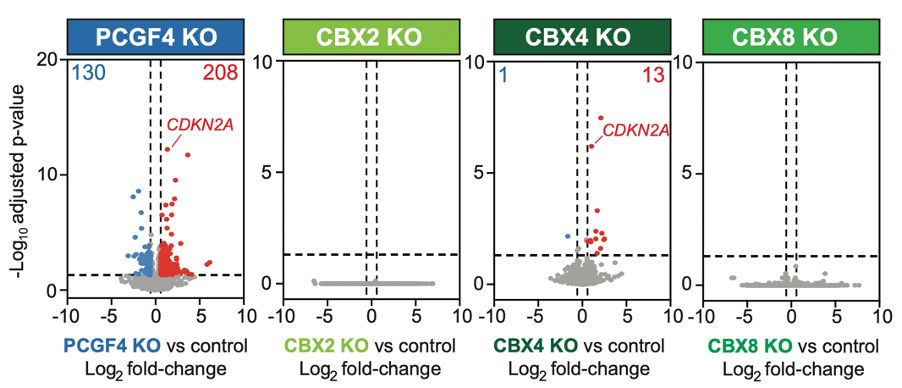
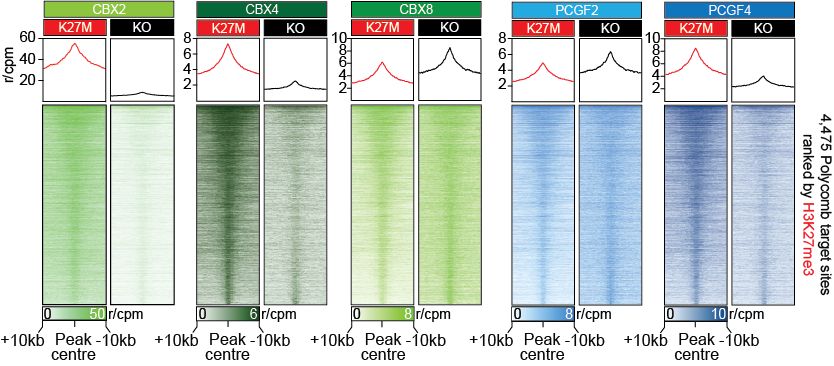
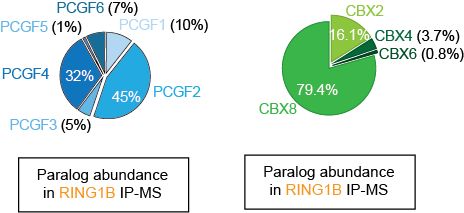
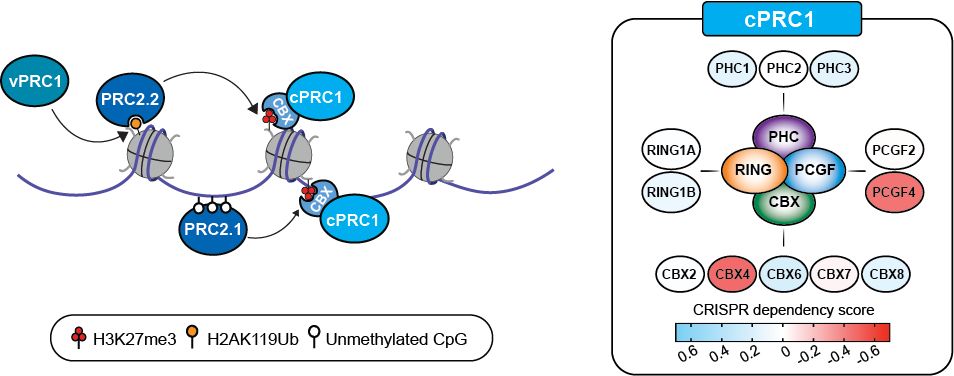
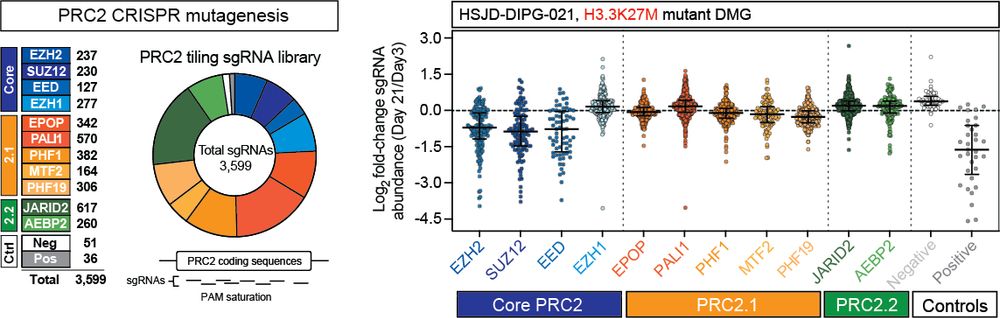
Lagan, Gannon, et al. reveal that H3K27M-DMGs depend on a specific form of cPRC1 containing CBX4 and PCGF4. H3K27M alters H3K27me3 distribution, causing increased binding of CBX4-PCGF4-cPRC1 and oncogenic gene repression. CBX4’s ability to read H3K27me3 and to form a specific cPRC1 complex with PCGF4 makes it essential in DMG.
dlvr.it
Eimear Lagan
@eimearlagan.bsky.social
· May 21
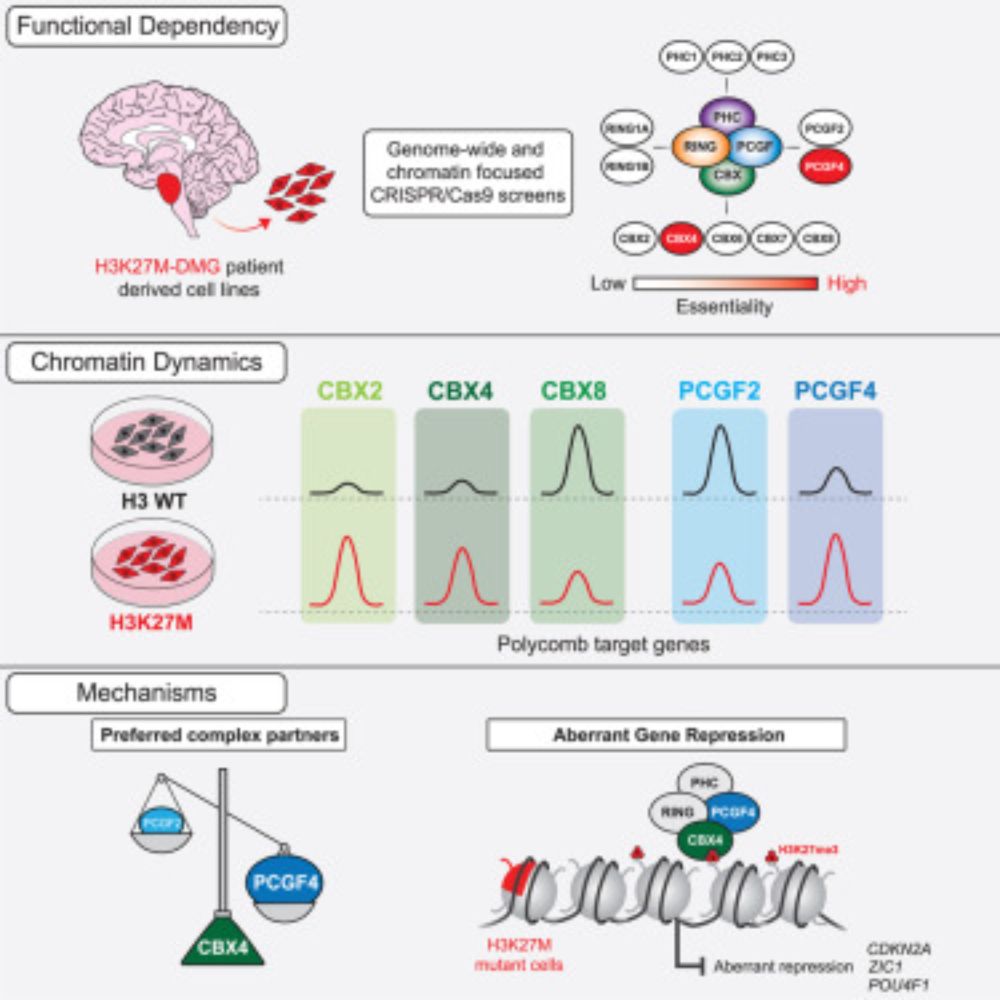
A specific form of cPRC1 containing CBX4 is co-opted to mediate oncogenic gene repression in diffuse midline glioma
Lagan, Gannon, et al. reveal that H3K27M-DMGs depend on a specific form of cPRC1 containing
CBX4 and PCGF4. H3K27M alters H3K27me3 distribution, causing increased binding of
CBX4-PCGF4-cPRC1 and oncog...
www.cell.com
Eimear Lagan
@eimearlagan.bsky.social
· May 21
Eimear Lagan
@eimearlagan.bsky.social
· May 21