Etowah Adams
@etowah0.bsky.social
88 followers
220 following
16 posts
enjoying and bemoaning biology. phd student
@columbia prev. @harvardmed @ginkgo @yale
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Etowah Adams
Tristan Bepler
@tbepler.bsky.social
· Feb 11
Reposted by Etowah Adams
Reposted by Etowah Adams
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10

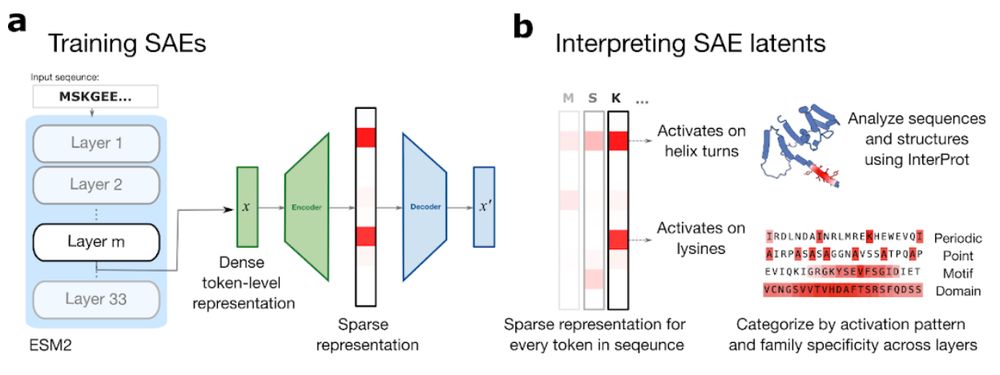
From Mechanistic Interpretability to Mechanistic Biology: Training, Evaluating, and Interpreting Sparse Autoencoders on Protein Language Models
Protein language models (pLMs) are powerful predictors of protein structure and function, learning through unsupervised training on millions of protein sequences. pLMs are thought to capture common mo...
www.biorxiv.org
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10
Etowah Adams
@etowah0.bsky.social
· Feb 10