Mohammed AlQuraishi
@moalquraishi.bsky.social
3.4K followers
63 following
24 posts
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.
Posts
Media
Videos
Starter Packs
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Nate Corley
@ncorley.bsky.social
· Aug 15

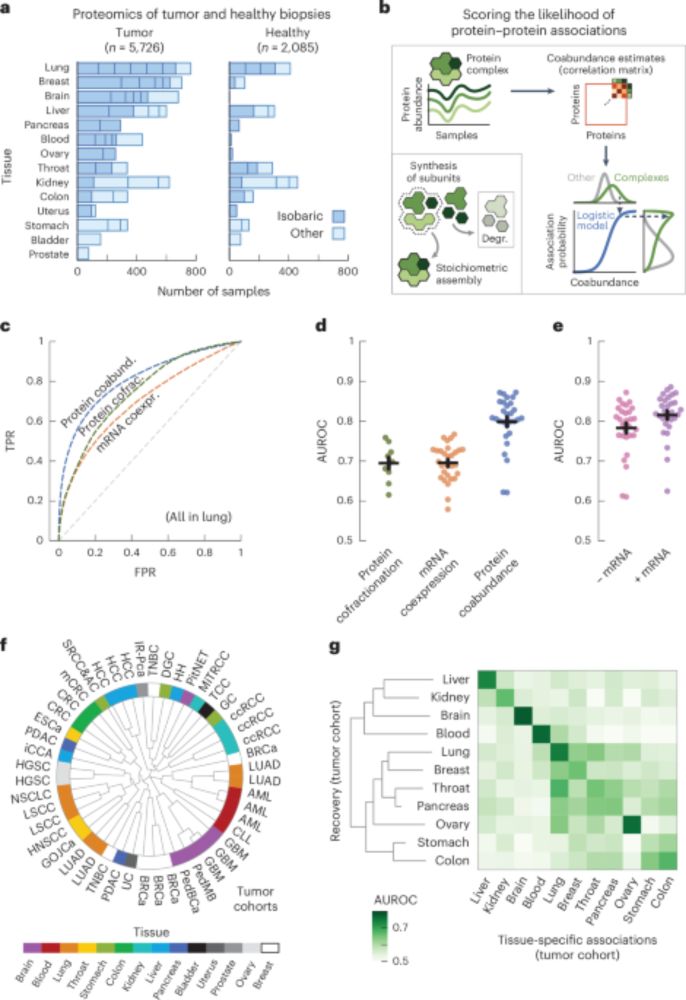
Accelerating Biomolecular Modeling with AtomWorks and RF3
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilita...
www.biorxiv.org
Reposted by Mohammed AlQuraishi
Tal Korem
@tkorem.bsky.social
· Aug 7
Korem Lab - Microbiome Systems Biology @ Columbia | New York, USA
We apply systems biology approaches to decipher the metabolic interactions between the microbiome and its human host in diverse clinical settings, aiming towards personalized microbiome-based therapeu...
www.koremlab.science
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Reposted by Mohammed AlQuraishi
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Feb 22

AlphaFold as a Prior: Experimental Structure Determination Conditioned on a Pretrained Neural Network
Advances in machine learning have transformed structural biology, enabling swift and accurate prediction of protein structure from sequence. However, challenges persist in capturing sidechain packing,...
www.biorxiv.org
Reposted by Mohammed AlQuraishi