Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
1.7K followers
160 following
61 posts
Interested in likelihood-based methods for structural biology. Contributor to Phenix, CCP4 and CCP-EM.
Based at CIMR, University of Cambridge.
Posts usually about science.
Posts
Media
Videos
Starter Packs
Arne Elofsson
@bioinfo.se
· 23d

Researcher offering insight into Parkinson’s awarded the Gregori Aminoff Prize - Kungl. Vetenskapsakademien
David Eisenberg has been awarded this year’s Gregori Aminoff Prize in Crystallography. He has pioneered the study of amyloid proteins’ molecular structure, providing new knowledge about neurodegenerat...
www.kva.se
Reposted by Randy Read 🇨🇦🇬🇧
Antoni Wrobel
@agwrobel.bsky.social
· Sep 8
Reposted by Randy Read 🇨🇦🇬🇧
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Aug 15
Reposted by Randy Read 🇨🇦🇬🇧
Reposted by Randy Read 🇨🇦🇬🇧
Titia Sixma
@titiasixma.bsky.social
· Jul 31
Niels Keijzer
@nkeijzer.bsky.social
· Jul 31
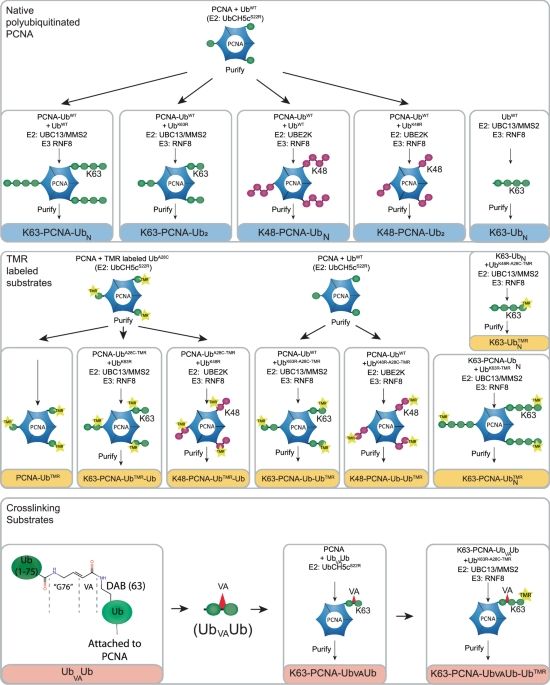
USP1/UAF1 targets polyubiquitinated PCNA with an exo-cleavage mechanism that can temporarily enrich for monoubiquitinated PCNA - Nature Communications
DNA damage tolerance is regulated by ubiquitination of PCNA. Here, the authors present kinetic and structural studies showing that USP1/UAF1 prefers trimming K63- and K48-ubiquitin chains down over cl...
doi.org
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Jul 29
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Jul 29
Reposted by Randy Read 🇨🇦🇬🇧
Structural Biology
@actacrystd.iucr.org
· Jul 29

Perspective on a large-scale ligand structure characterization
An introduction to a set of three related papers in this issue describing 216 ligand-bound fatty acid-binding protein structure determinations and the pitfalls that can arise in such a study.
journals.iucr.org
Reposted by Randy Read 🇨🇦🇬🇧
Alexandre Bonvin
@amjjbonvin.bsky.social
· Jul 29
Reposted by Randy Read 🇨🇦🇬🇧
Reposted by Randy Read 🇨🇦🇬🇧
IUCr2026
@iucr2026.bsky.social
· Jul 28
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Jul 24
IUCr2026
@iucr2026.bsky.social
· Jul 24

RCSB PDB - 3SGB: STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION
STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION
www.rcsb.org
Randy Read 🇨🇦🇬🇧
@randyjread.bsky.social
· Jul 24
Miro Astore
@miroastore.bsky.social
· Jul 23

The Inaugural Flatiron Institute Cryo-EM Conformational Heterogeneity Challenge
Despite the rise of single particle cryo-electron microscopy (cryo-EM) as a premier method for resolving macromolecular structures at atomic resolution, methods to address molecular heterogeneity in v...
www.biorxiv.org







