Oli Clarke
@olibclarke.bsky.social
1.1K followers
710 following
360 posts
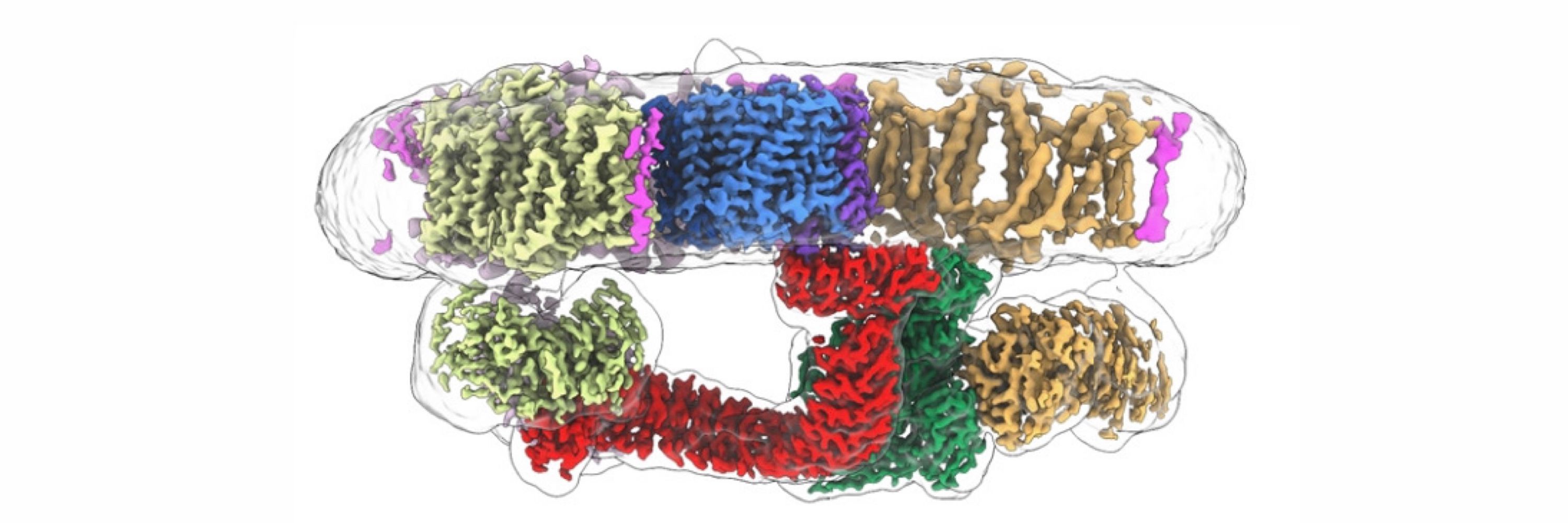
CryoEM, membrane proteins and whatnot
Posts
Media
Videos
Starter Packs
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke
Reposted by Oli Clarke