Jan-Hannes Schaefer
@jhschaef.bsky.social
380 followers
410 following
54 posts
Structural Biology to the rescue. Postdoc with Gabriel Lander at Scripps Research. PhD with Arne Moeller. #cryoEM ❄️🔬 🎹⛳
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jan-Hannes Schaefer
Reposted by Jan-Hannes Schaefer
Reposted by Jan-Hannes Schaefer
Reposted by Jan-Hannes Schaefer
Reposted by Jan-Hannes Schaefer
Jan-Hannes Schaefer
@jhschaef.bsky.social
· Aug 31
Jan-Hannes Schaefer
@jhschaef.bsky.social
· Aug 29
Reposted by Jan-Hannes Schaefer
Reposted by Jan-Hannes Schaefer
David B. Sauer
@davidbsauer.bsky.social
· Aug 15

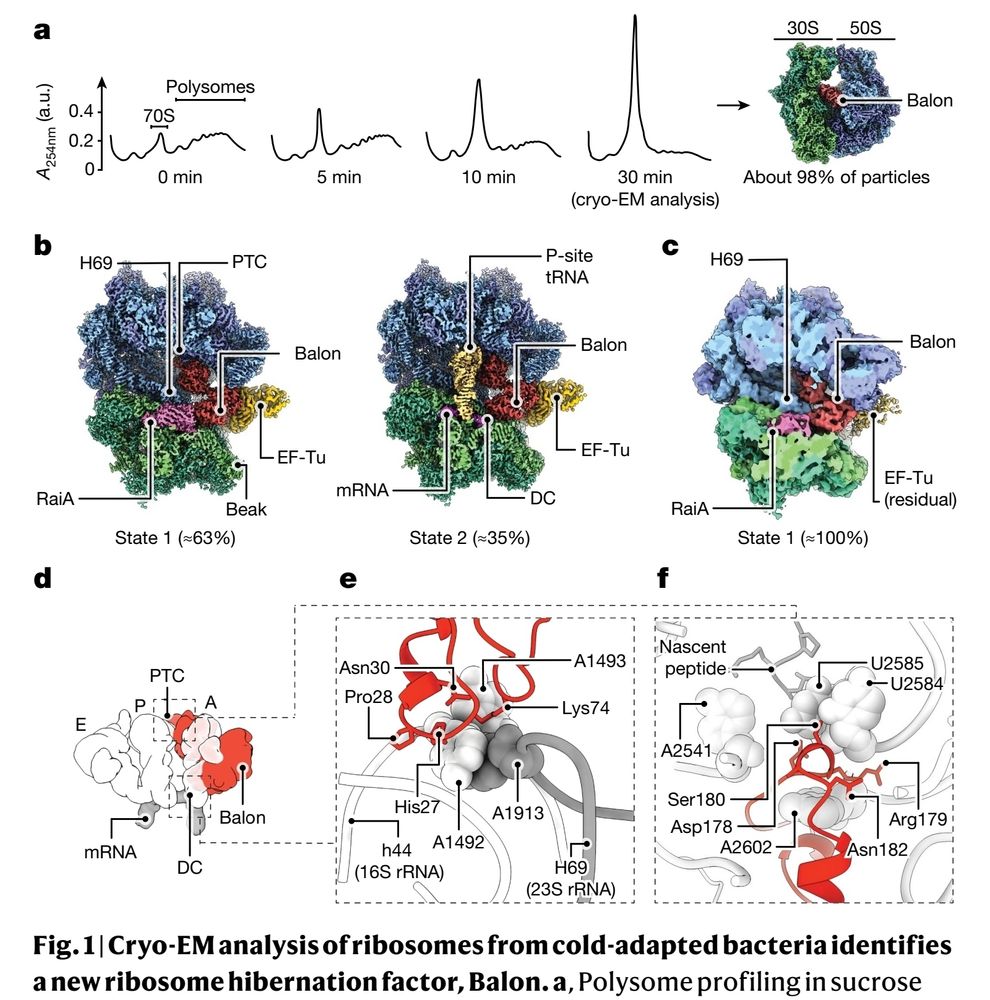
Covalently constrained ‘Di-Gembodies’ enable parallel structure solutions by cryo-EM - Nature Chemical Biology
Disulfide-based dimerization of modified identical and heterologous nanobody scaffolds enables higher-order assembly for high-resolution cryo-electron microscopy structure determination that is widely...
www.nature.com
Jan-Hannes Schaefer
@jhschaef.bsky.social
· Aug 14
Jan-Hannes Schaefer
@jhschaef.bsky.social
· Aug 14
Reposted by Jan-Hannes Schaefer
Eric Topol
@erictopol.bsky.social
· Aug 6
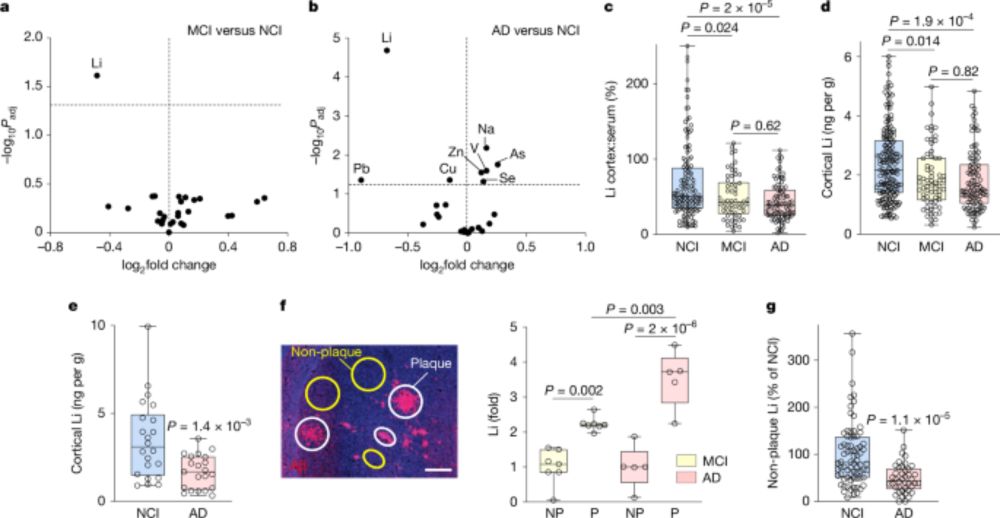
Lithium deficiency and the onset of Alzheimer’s disease - Nature
Lithium has an essential role in the brain and is deficient early in Alzheimer’s disease, which can be recapitulated in mice and treated with a novel lithium salt that restores the physiological level.
nature.com