Julia Rogers
@juliarurogers.bsky.social
330 followers
550 following
12 posts
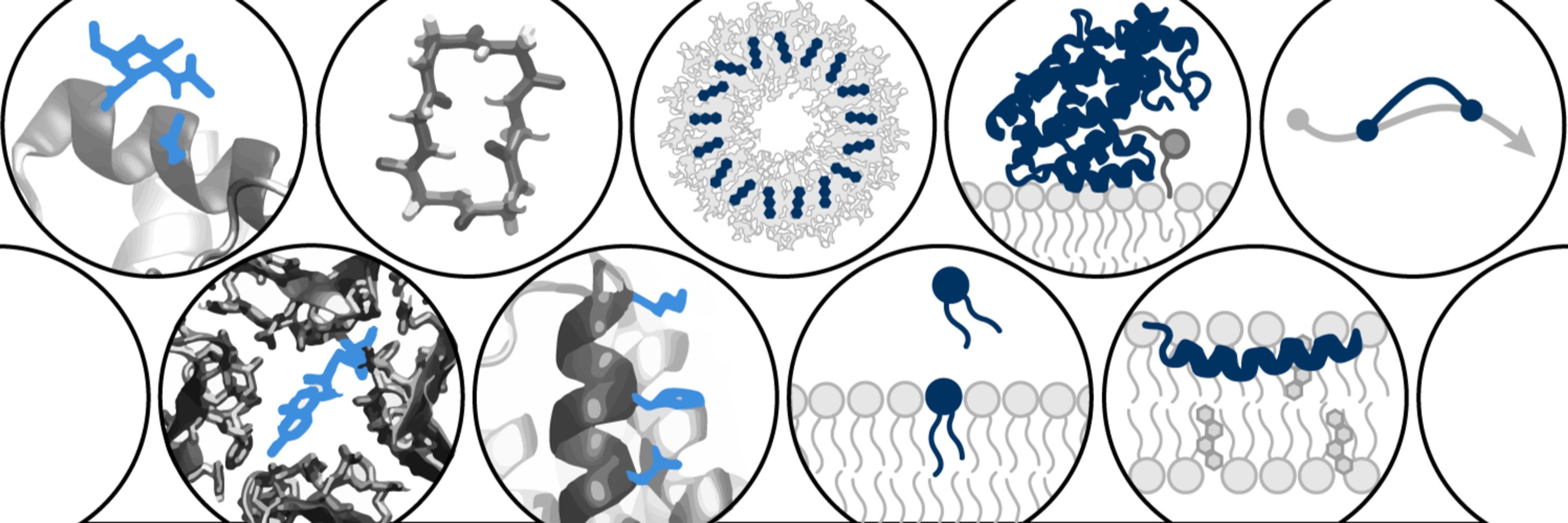
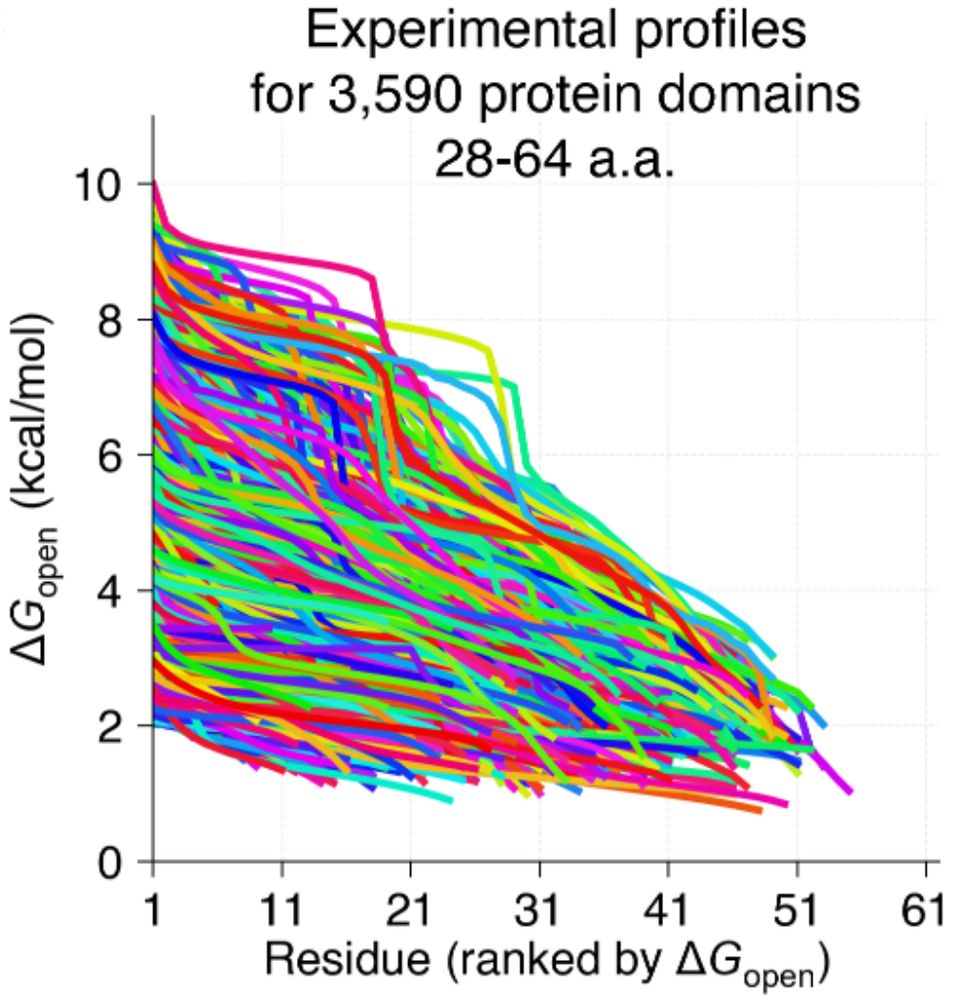
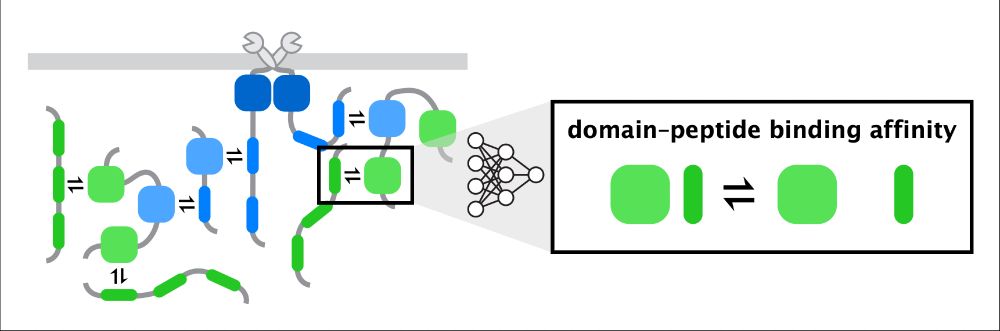
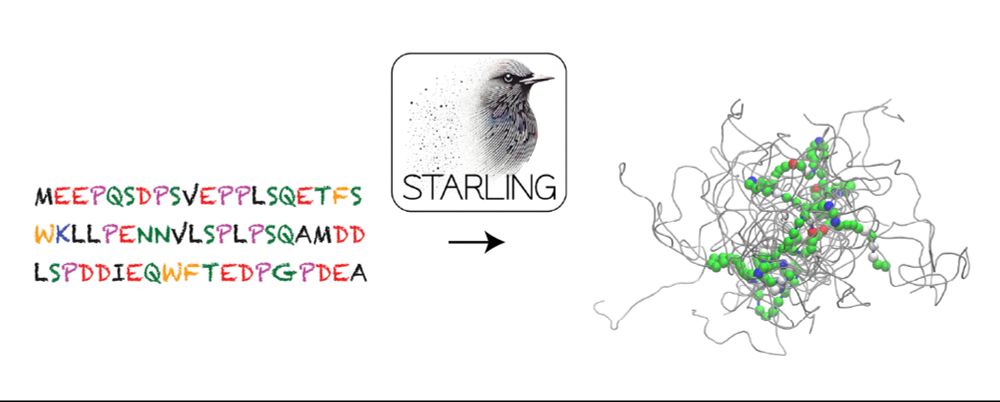
BWF CASI Fellow @Columbia | 2022 Jane Coffin Childs Fellow | PhD @UCBerkeley | BS @TuftsUniversity | Systems biophysics via integrative ML- and physics-based models
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Julia Rogers
Julia Rogers
@juliarurogers.bsky.social
· Apr 13
Reposted by Julia Rogers
Julia Rogers
@juliarurogers.bsky.social
· Mar 26
Reposted by Julia Rogers
Reposted by Julia Rogers
Reposted by Julia Rogers
Reposted by Julia Rogers
Reposted by Julia Rogers
Alice Ting
@aliceyting.bsky.social
· Dec 4
Reposted by Julia Rogers