Marek Glombik
@glombikm.bsky.social
600 followers
450 following
2 posts
postdoc in De Vega lab, Earlham Institute, Norwich, UK.
Pangenomes, polyploid crops, evolution and improvement.
Posts
Media
Videos
Starter Packs
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
K.D. Murray
@kdm9.bsky.social
· Aug 14
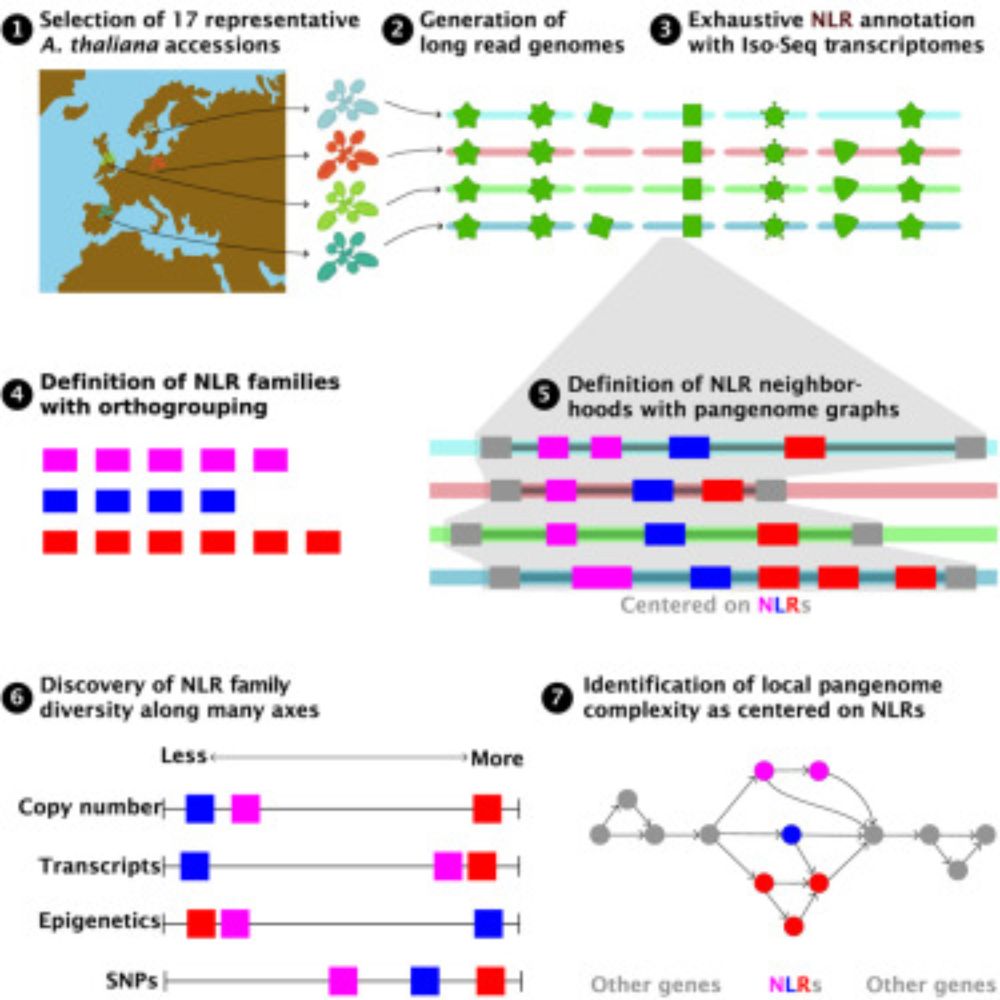
Pangenomic context reveals the extent of intraspecific plant NLR evolution
Individual- and population-level diversity is required for pathogen defense by nucleotide-binding
site leucine-rich repeat (NLR) proteins. Teasdale et al. leverage annotated, divergent
A. thaliana gen...
www.cell.com
Reposted by Marek Glombik
Stephen Turner
@stephenturner.us
· Aug 14
Prompt-based bioinformatics: a new interface for multi-omics analysis - Nature Reviews Genetics
Prompt-based methods, which involve the careful design of inputs to guide large language model (LLM) outputs, are beginning to reshape bioinformatic analytical workflows. The authors compare prompt-dr...
www.nature.com
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Facundo Romani
@fromani.bsky.social
· Jul 21

Conservation of chromatin states and their association with transcription factors in land plants
The complexity of varied modifications of chromatin composition is integrated in archetypal combinations called chromatin states that predict the local potential for transcription. The degree of conse...
www.biorxiv.org
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik
Reposted by Marek Glombik