Posts
Media
Videos
Starter Packs
Reposted by Alexis Caulier
Alexis Caulier
@hemagene.bsky.social
· Apr 16

A genome-wide screen identifies genes required for erythroid differentiation - Nature Communications
Using a genome-wide CRISPR knock-out screen, the authors defined the repertoire of genes that are required for proerythroblast survival and for terminal erythroid differentiation.
www.nature.com
Alexis Caulier
@hemagene.bsky.social
· Apr 5
Reposted by Alexis Caulier
Alexis Caulier
@hemagene.bsky.social
· Apr 3
Alexis Caulier
@hemagene.bsky.social
· Apr 3
Alexis Caulier
@hemagene.bsky.social
· Apr 3
Alexis Caulier
@hemagene.bsky.social
· Apr 3
Alexis Caulier
@hemagene.bsky.social
· Apr 3
Alexis Caulier
@hemagene.bsky.social
· Apr 3
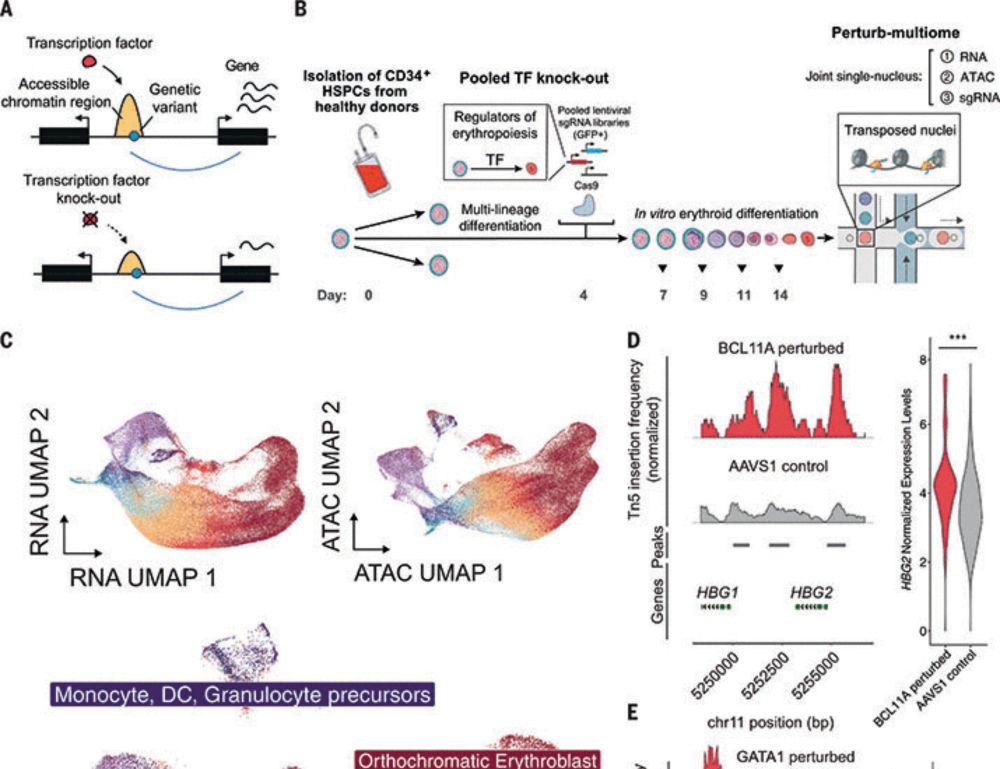
Transcription factor networks disproportionately enrich for heritability of blood cell phenotypes
Most phenotype-associated genetic variants map to noncoding regulatory regions of the human genome, but their mechanisms remain elusive in most cases. We developed a highly efficient strategy, Perturb...
bit.ly
Reposted by Alexis Caulier












