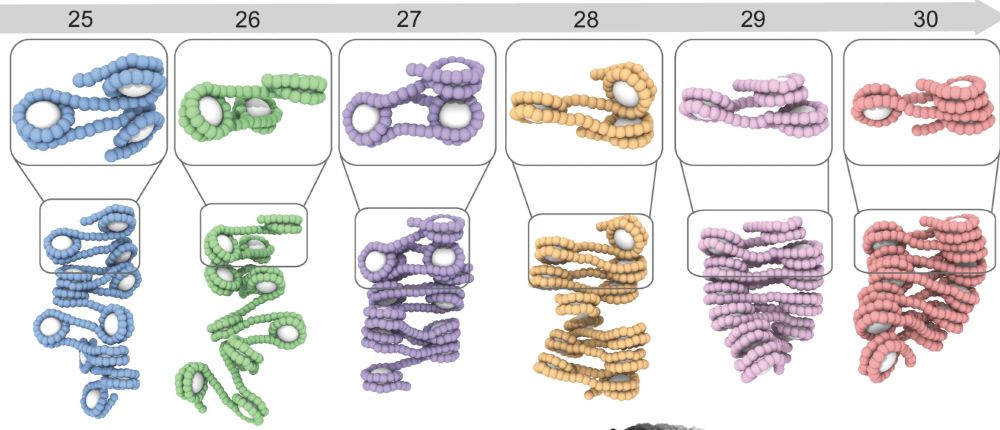
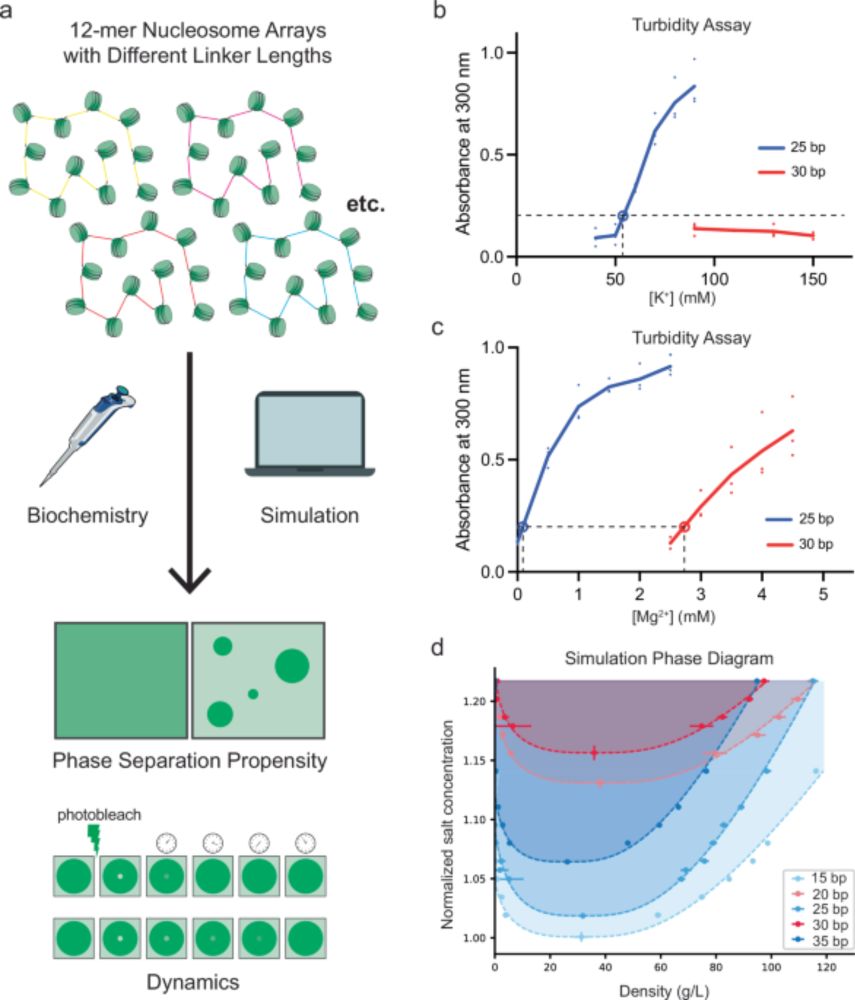
Julia Maristany
@juliamaristany.bsky.social
120 followers
120 following
10 posts
Postdoctoral Researcher at the Collepardo Lab, in Cambridge, working on chromatin phase separation
She/Her :) A science, dog, photography person, and most of all, an absolute chromatin geek
Posts
Media
Videos
Starter Packs
Reposted by Julia Maristany
Reposted by Julia Maristany
Miguel Montez
@miguelmontez.bsky.social
· Feb 21

Cold-induced nucleosome dynamics linked to silencing of Arabidopsis FLC
Temperature influences nucleosome dynamics, and thus chromatin, to regulate gene expression. Such mechanisms underlie the epigenetic silencing of Arabidopsis FLOWERING LOCUS C (FLC) by prolonged cold....
www.biorxiv.org
Reposted by Julia Maristany
Reposted by Julia Maristany
Reposted by Julia Maristany