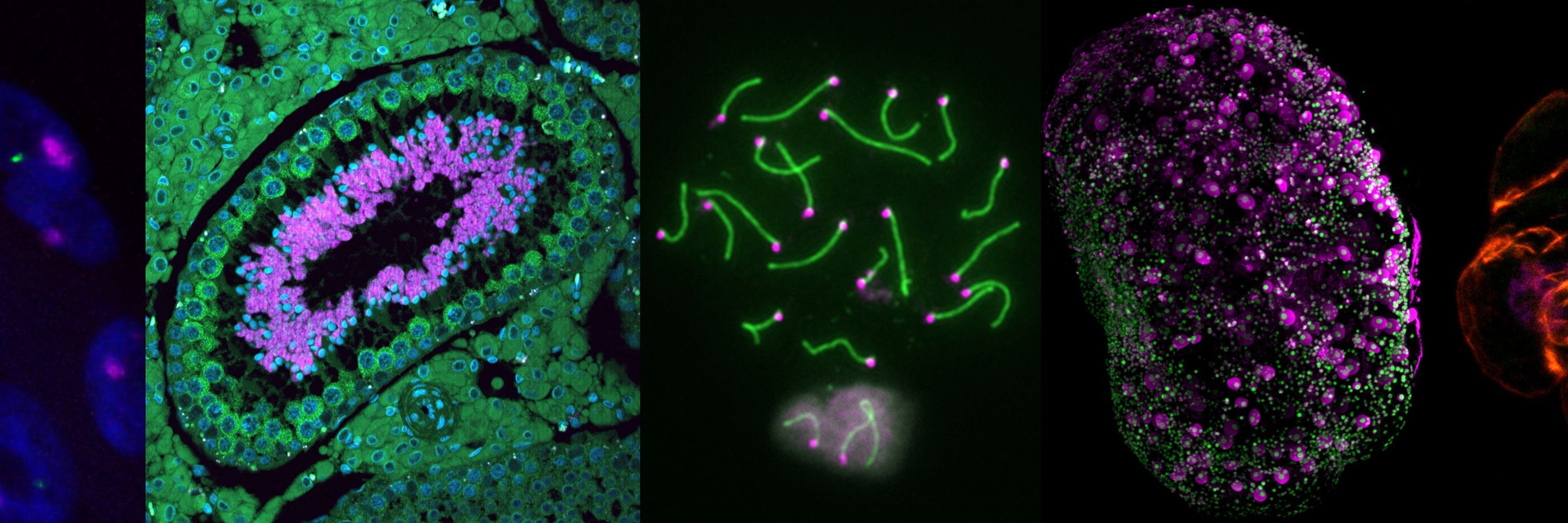
Turner Lab
@lab-turner.bsky.social
300 followers
210 following
23 posts
Account of the Turner lab at the Francis Crick Institute in London. We study sex chromosomes and their impact on health and disease. Rotating curation by lab members.
https://www.crick.ac.uk/research/labs/james-turner
Posts
Media
Videos
Starter Packs
Pinned
Turner Lab
@lab-turner.bsky.social
· May 14
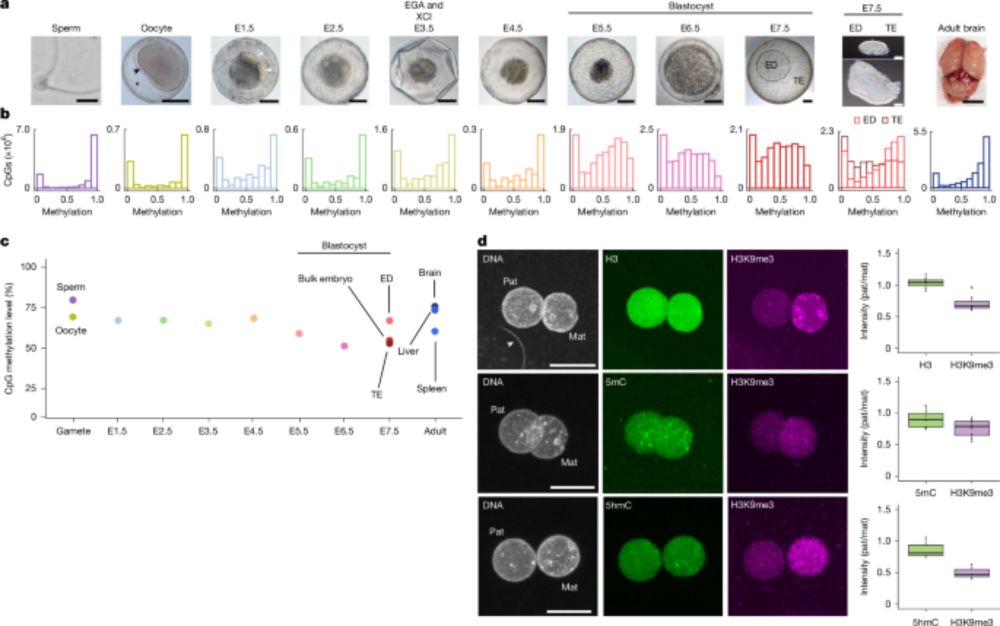
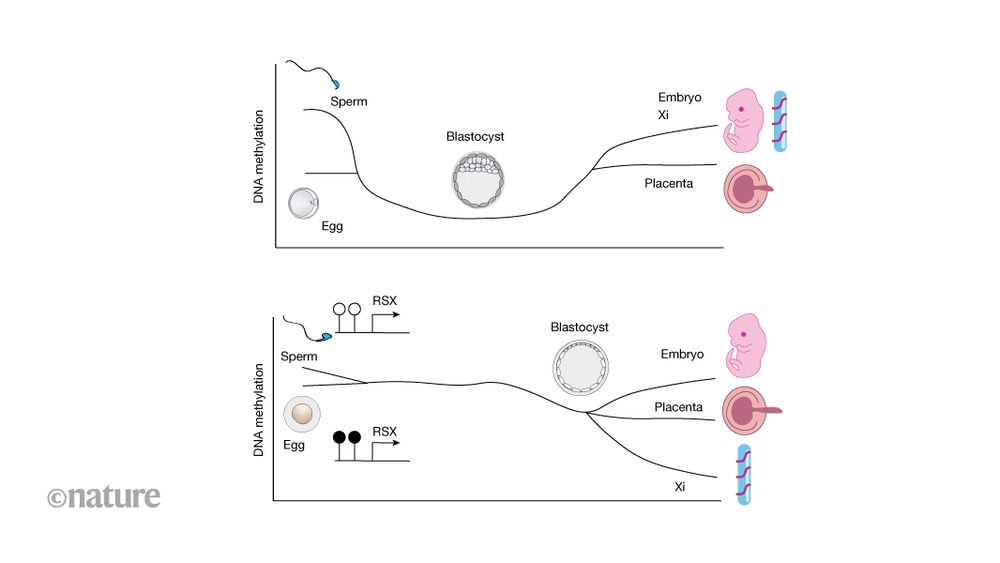
Divergent DNA methylation dynamics in marsupial and eutherian embryos - Nature
A study reports on the DNA methylation dynamics during embryogenesis in marsupials, showing that these differ from those occurring during embryogenesis in eutherian mammals.
www.nature.com
Reposted by Turner Lab
Sergio Menchero
@sermenchero.bsky.social
· Aug 11
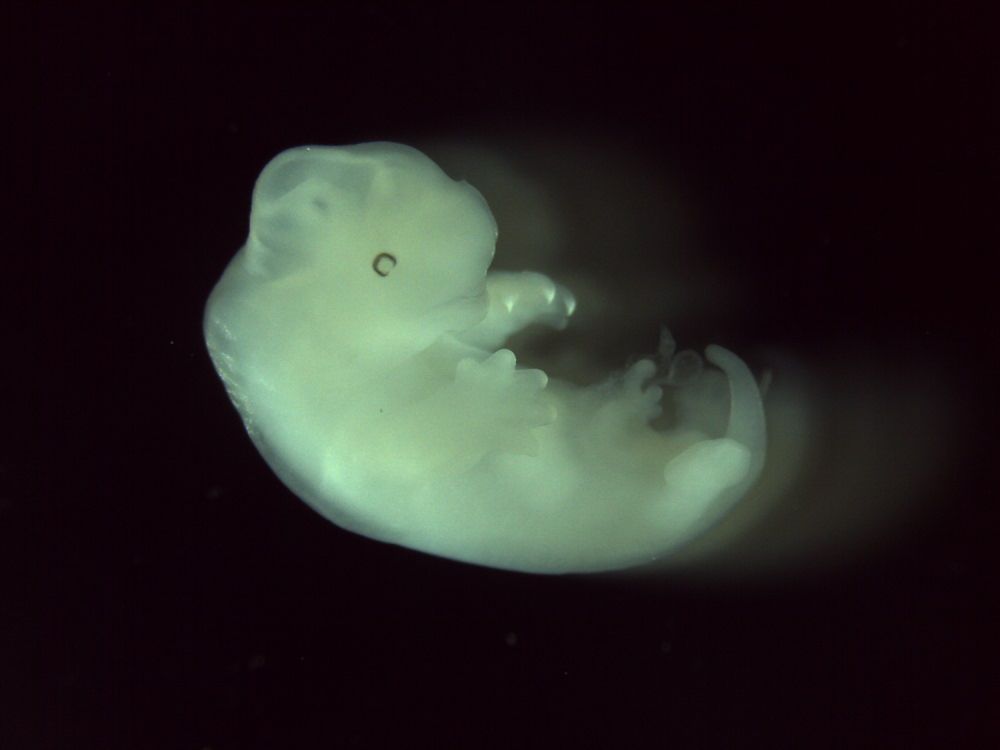
In opossums, gene expression follows familiar rules but at a strange pace
The arms and heads of opossums (pictured here one day before birth) and other marsupials develop faster than their legs and back bodies. Image credit: Sergio Menchero Fernandez/ Francis Crick Institut...
www.pnas.org
Turner Lab
@lab-turner.bsky.social
· Jul 25
Sergio Menchero
@sermenchero.bsky.social
· Jul 24
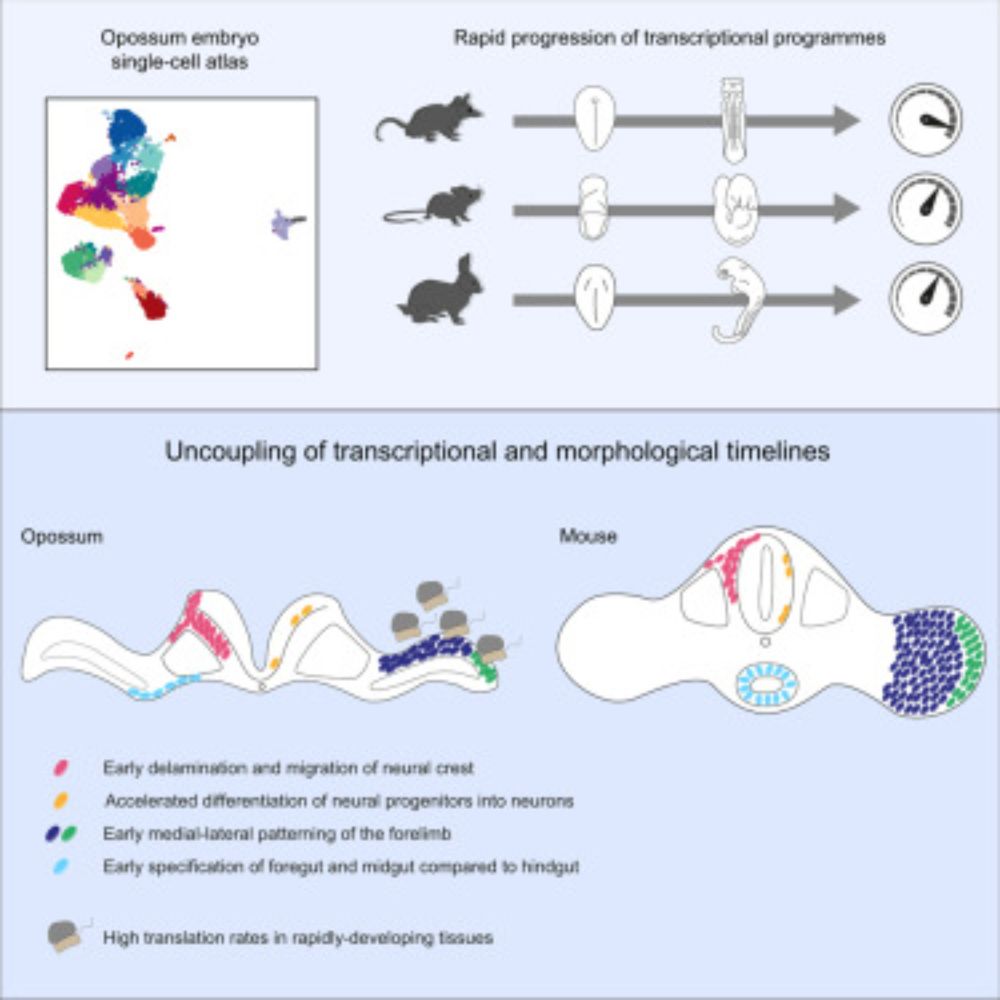
Marsupial single-cell transcriptomics identifies temporal diversity in mammalian developmental programs
Menchero et al. generate a single-cell transcriptomic atlas in the opossum and show
rapid progression of transcriptional programs in specific tissues relative to morphological
landmarks. This shift in...
www.cell.com
Reposted by Turner Lab
Turner Lab
@lab-turner.bsky.social
· May 15
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14
Turner Lab
@lab-turner.bsky.social
· May 14

Divergent DNA methylation dynamics in marsupial and eutherian embryos - Nature
A study reports on the DNA methylation dynamics during embryogenesis in marsupials, showing that these differ from those occurring during embryogenesis in eutherian mammals.
www.nature.com
Turner Lab
@lab-turner.bsky.social
· Mar 7
Turner Lab
@lab-turner.bsky.social
· Mar 7

A human induced pluripotent stem cell toolbox for studying sex chromosome effects
Sex chromosomes shape male (XY) - female (XX) differences in development and disease. These differences can be modelled in vitro by comparing XY and XX human induced pluripotent stem cells (hiPSCs). H...
www.biorxiv.org
Reposted by Turner Lab
Turner Lab
@lab-turner.bsky.social
· Jan 27
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24
Turner Lab
@lab-turner.bsky.social
· Jan 24