Manu Saraswat
@manusaraswat.bsky.social
230 followers
200 following
39 posts
PhD candidate in ML for genomics in Heidelberg, Germany with Oli Stegle
Previously at Genentech, UBC and BITS Pilani
https://scholar.google.com/citations?user=4yUtALcAAAAJ&hl=en&oi=ao
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Manu Saraswat
Reposted by Manu Saraswat
Peter Koo
@pkoo562.bsky.social
· Jul 16
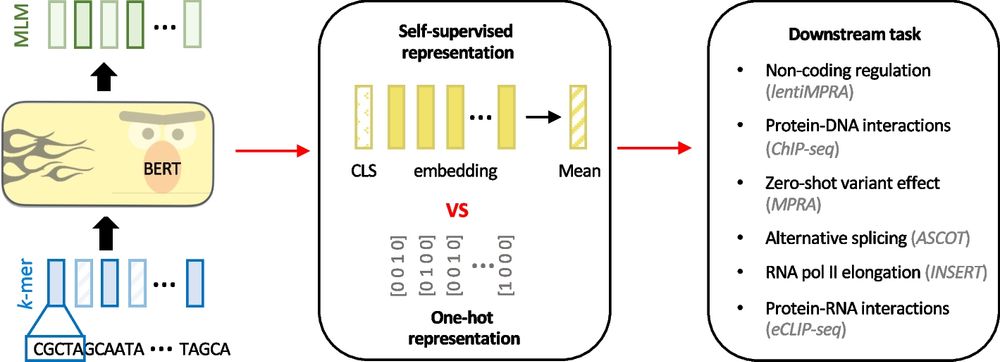
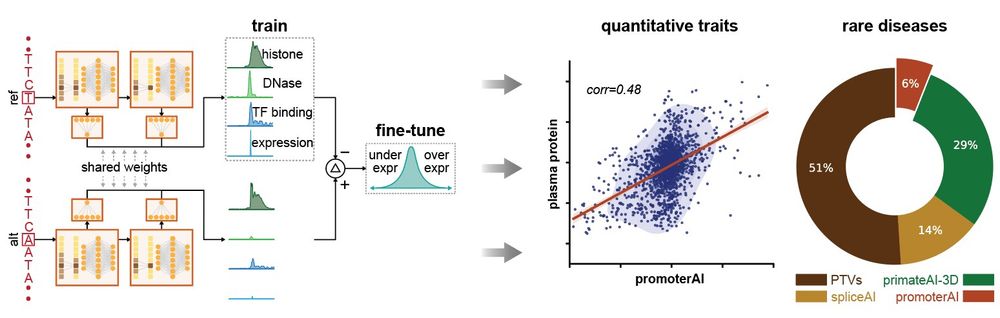
Evaluating the representational power of pre-trained DNA language models for regulatory genomics - Genome Biology
Background The emergence of genomic language models (gLMs) offers an unsupervised approach to learning a wide diversity of cis-regulatory patterns in the non-coding genome without requiring labels of ...
genomebiology.biomedcentral.com
Manu Saraswat
@manusaraswat.bsky.social
· Jun 23
Reposted by Manu Saraswat
Manu Saraswat
@manusaraswat.bsky.social
· May 22
Manu Saraswat
@manusaraswat.bsky.social
· May 22
Manu Saraswat
@manusaraswat.bsky.social
· May 22
Reposted by Manu Saraswat
Manu Saraswat
@manusaraswat.bsky.social
· May 16
Manu Saraswat
@manusaraswat.bsky.social
· May 16
Manu Saraswat
@manusaraswat.bsky.social
· May 16
Manu Saraswat
@manusaraswat.bsky.social
· May 16

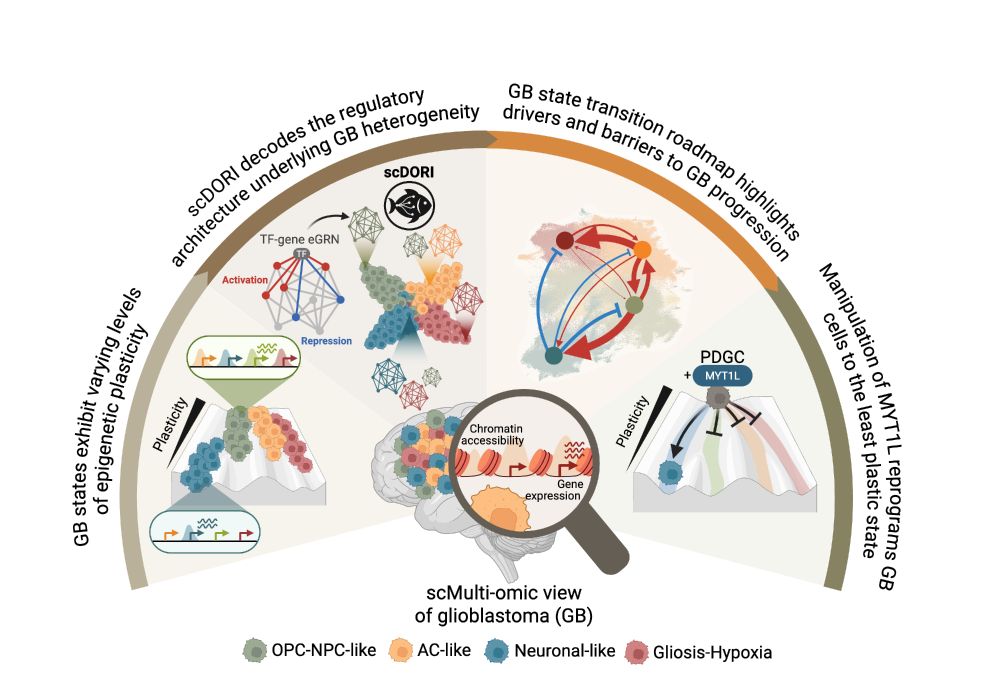
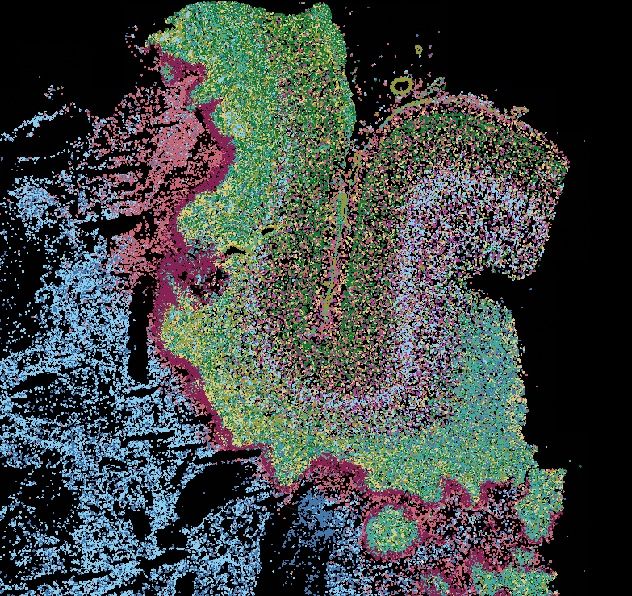
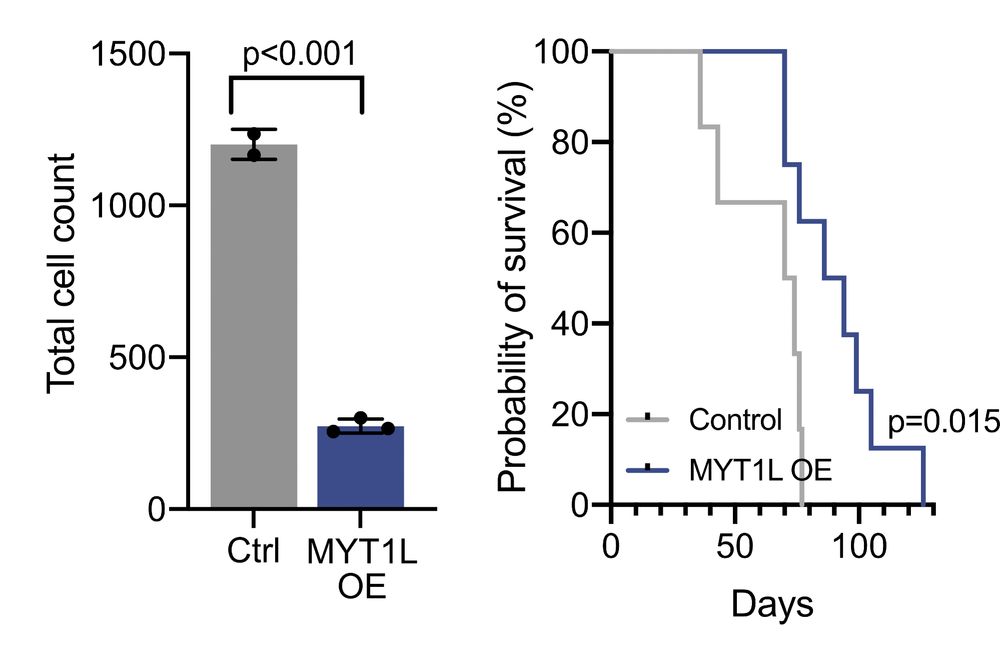
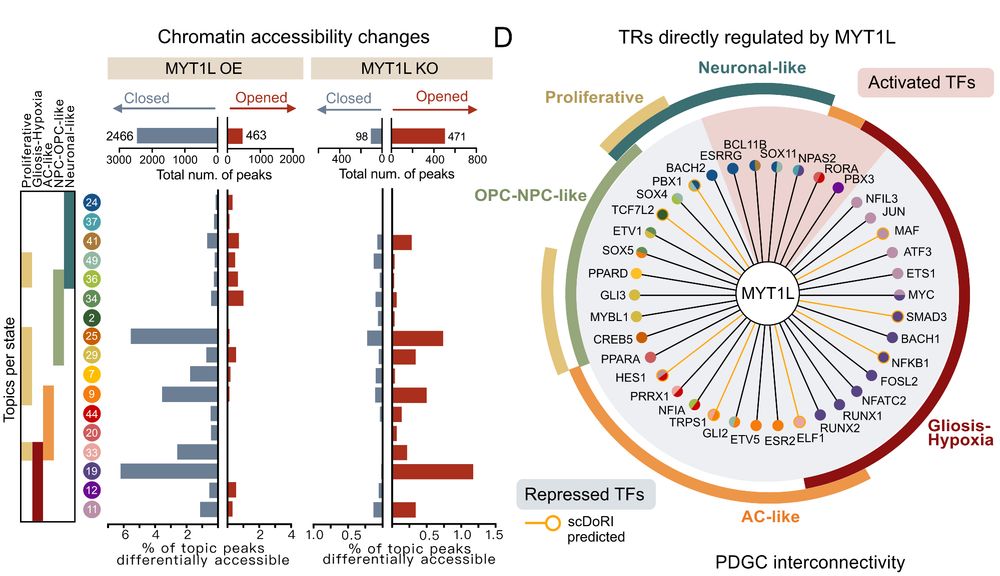
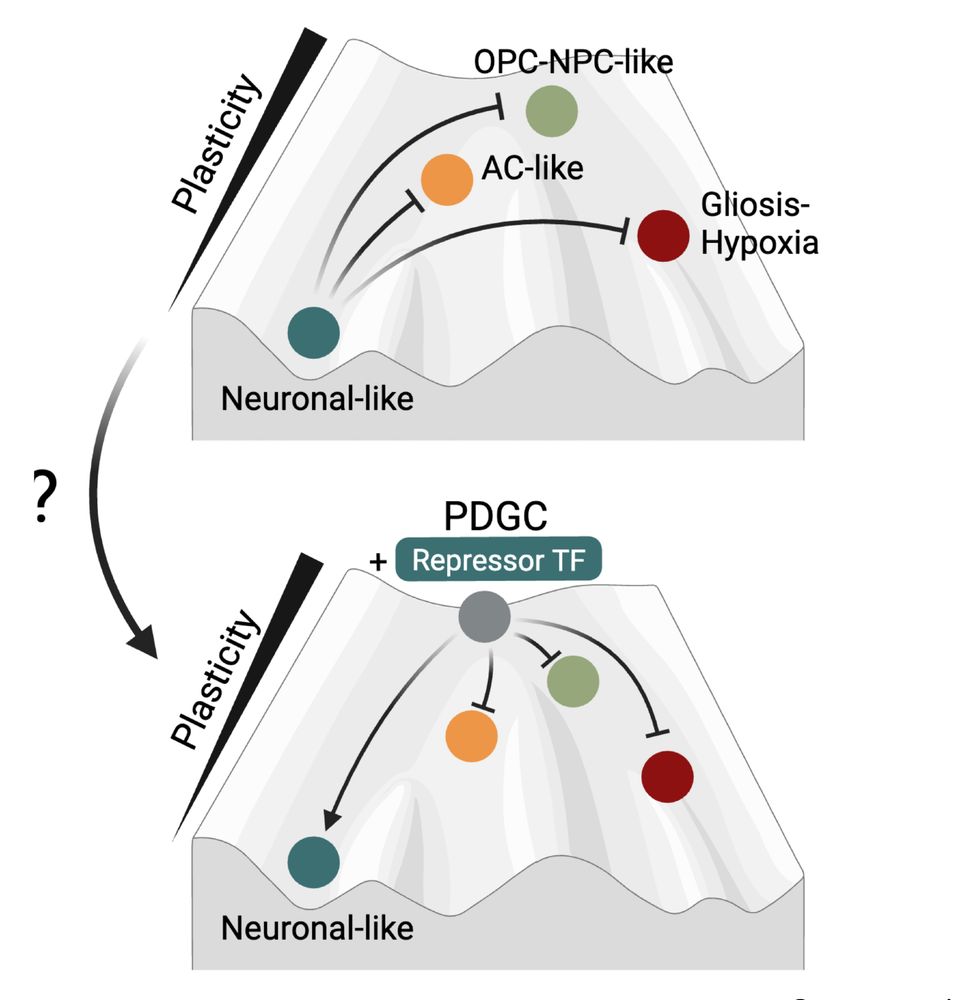
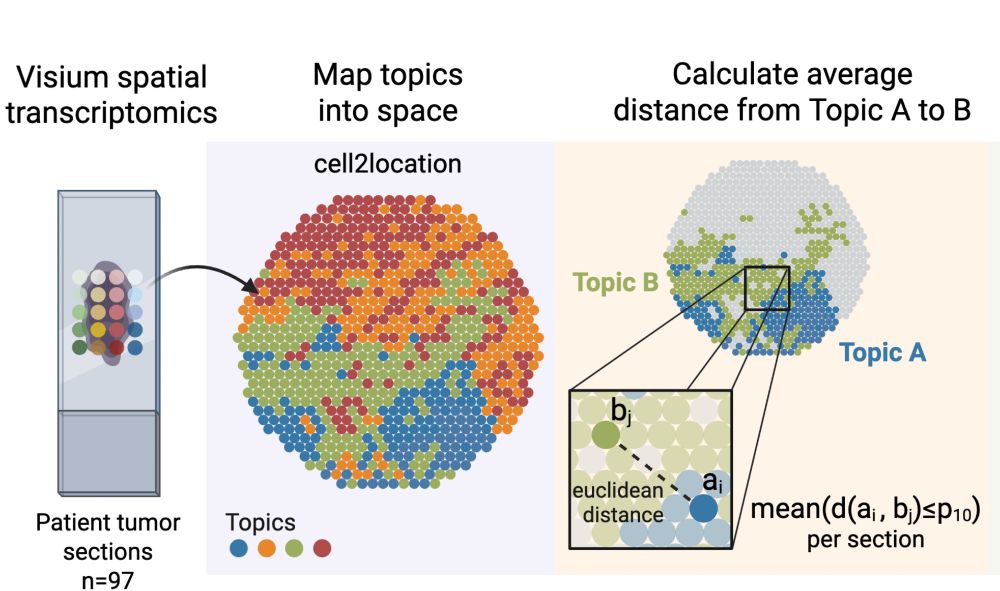
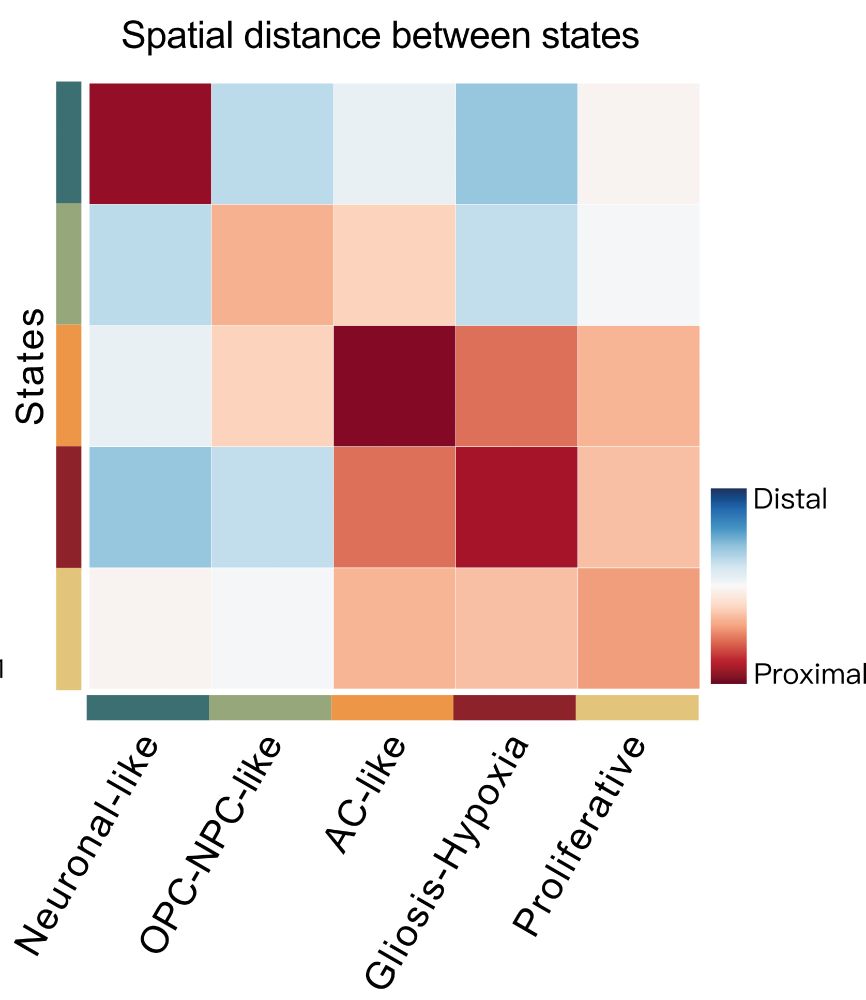
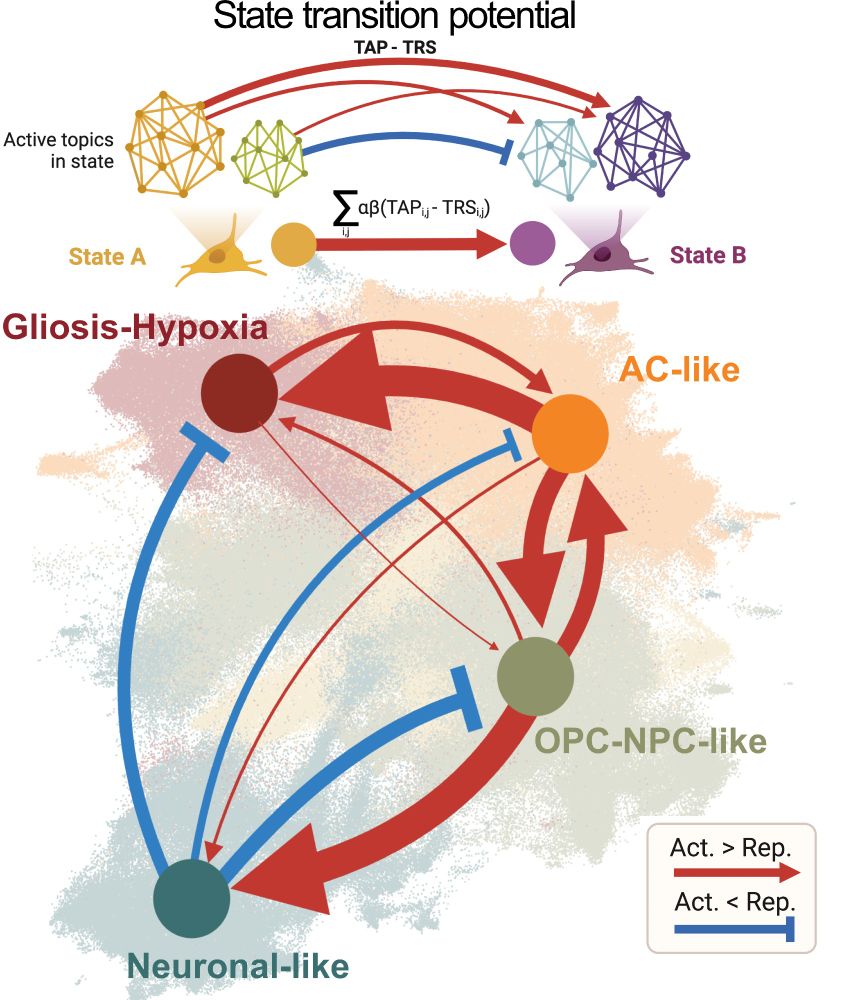
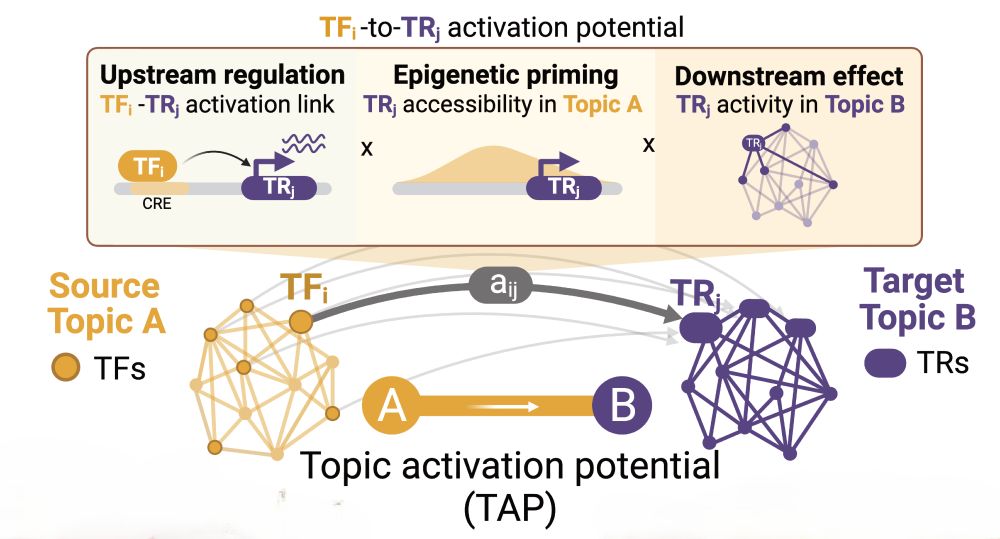
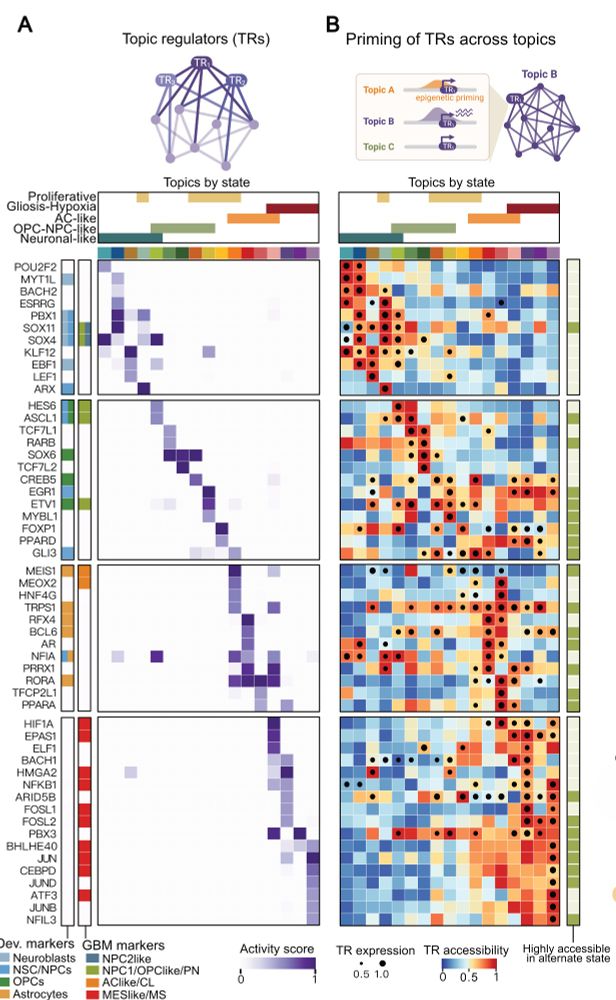
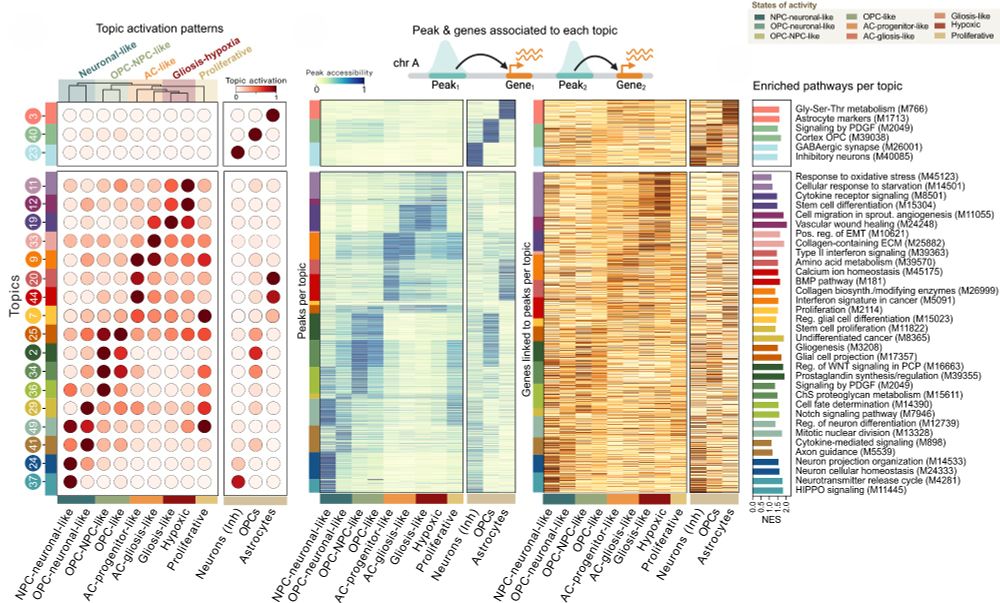
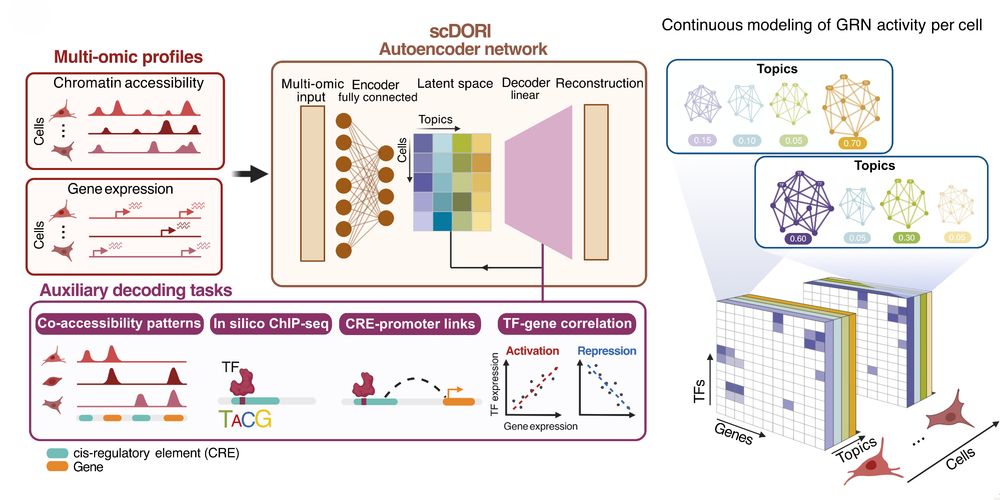
Decoding Plasticity Regulators and Transition Trajectories in Glioblastoma with Single-cell Multiomics
Glioblastoma (GB) is one of the most lethal human cancers, marked by profound intratumoral heterogeneity and near-universal treatment resistance. Cellular plasticity, the capacity of cancer cells to t...
www.biorxiv.org
Manu Saraswat
@manusaraswat.bsky.social
· May 16
Manu Saraswat
@manusaraswat.bsky.social
· May 16