Omer Ali Bayraktar
@bayraktarlab.bsky.social
210 followers
210 following
27 posts
Group leader
@sangerinstitute.bsky.social Neural diversity, spatial transcriptomics, glia, GBM
Posts
Media
Videos
Starter Packs
Reposted by Omer Ali Bayraktar
Open Targets
@opentargets.org
· Jul 3
Technical Specialist: Neurodegenerative Disease Spatial Transcriptomics and Data Generation
Do you want to help us improve human health and understand life on Earth? Make your mark by shaping the future to enable or deliver life-changing science to solve some of humanity’s greatest challenge...
sanger.wd103.myworkdayjobs.com
Reposted by Omer Ali Bayraktar
Open Targets
@opentargets.org
· Jul 3
Postdoctoral Fellow: Neurodegenerative Disease Spatial Transcriptomics and Machine Learning
Do you want to help us improve human health and understand life on Earth? Make your mark by shaping the future to enable or deliver life-changing science to solve some of humanity’s greatest challenge...
sanger.wd103.myworkdayjobs.com
Reposted by Omer Ali Bayraktar
Jimmy Lee
@drjimmylee.bsky.social
· Jun 30
Reposted by Omer Ali Bayraktar
Stein Aerts
@steinaerts.bsky.social
· May 16

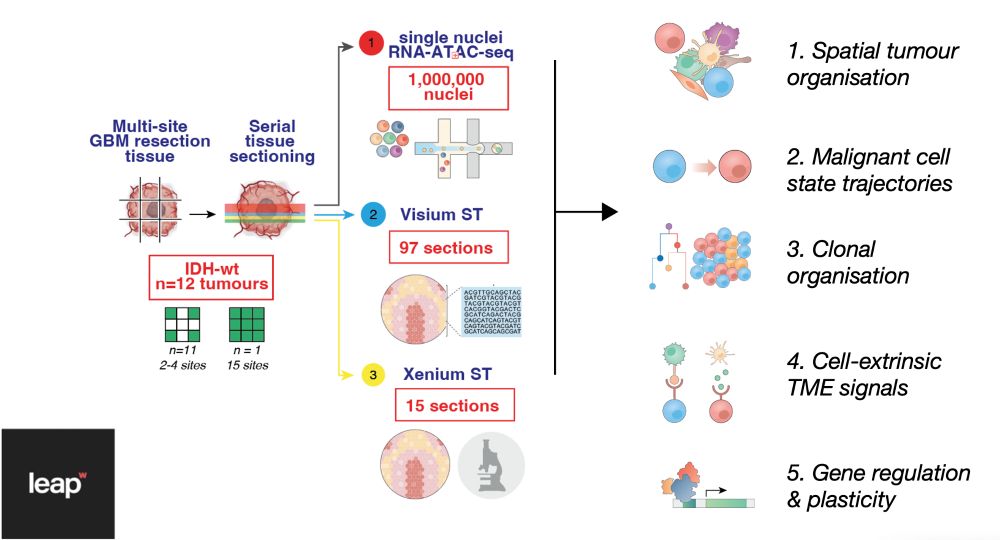
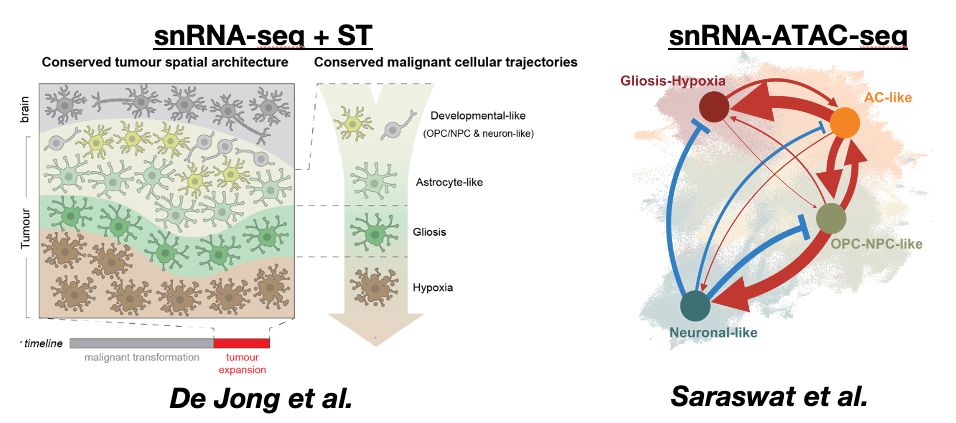
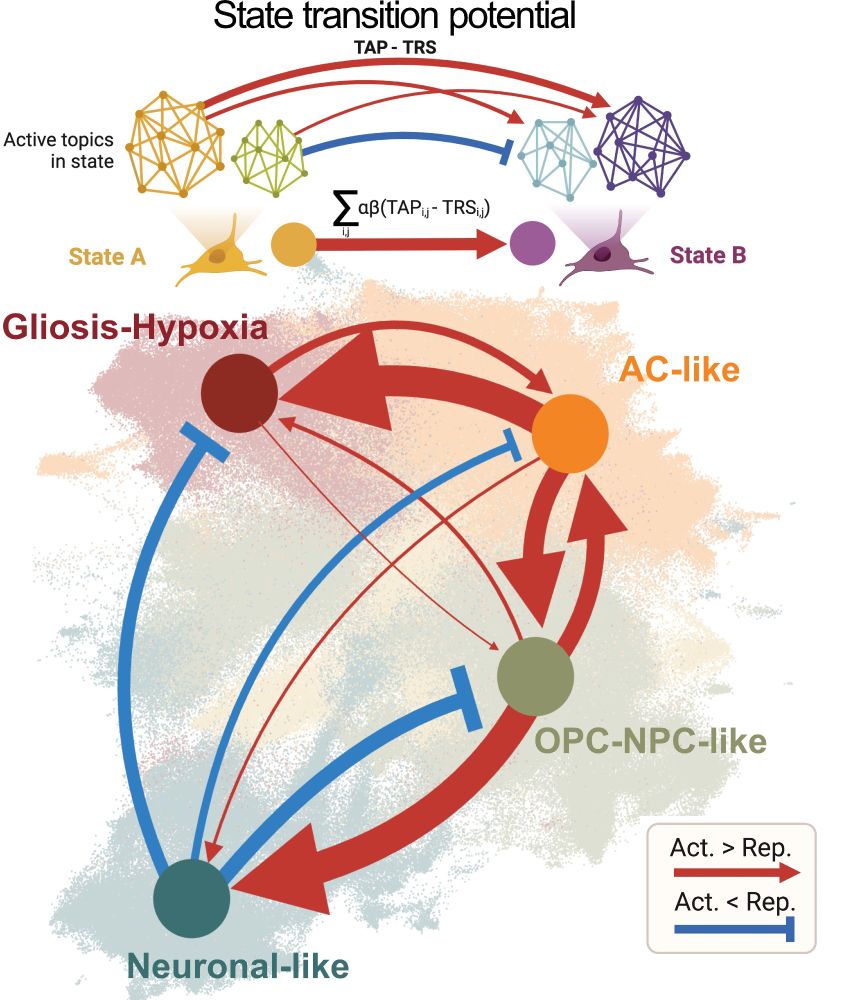
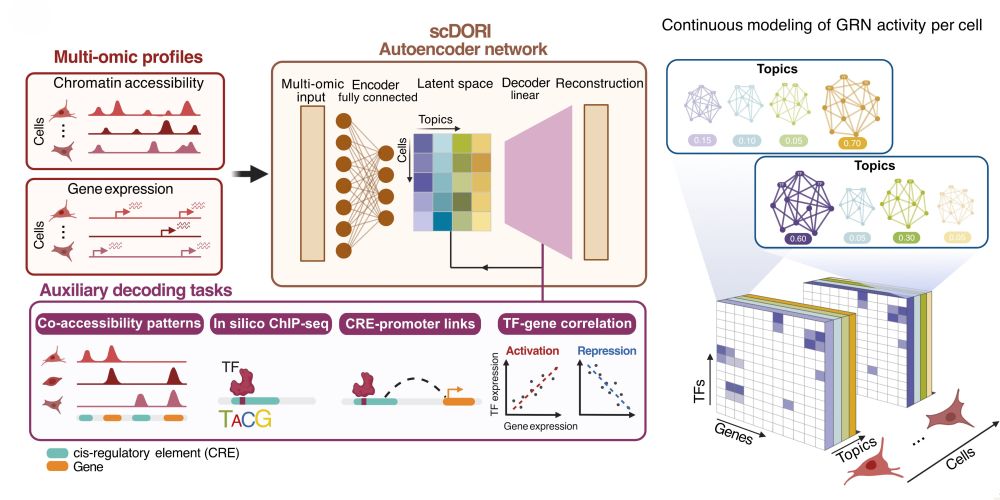
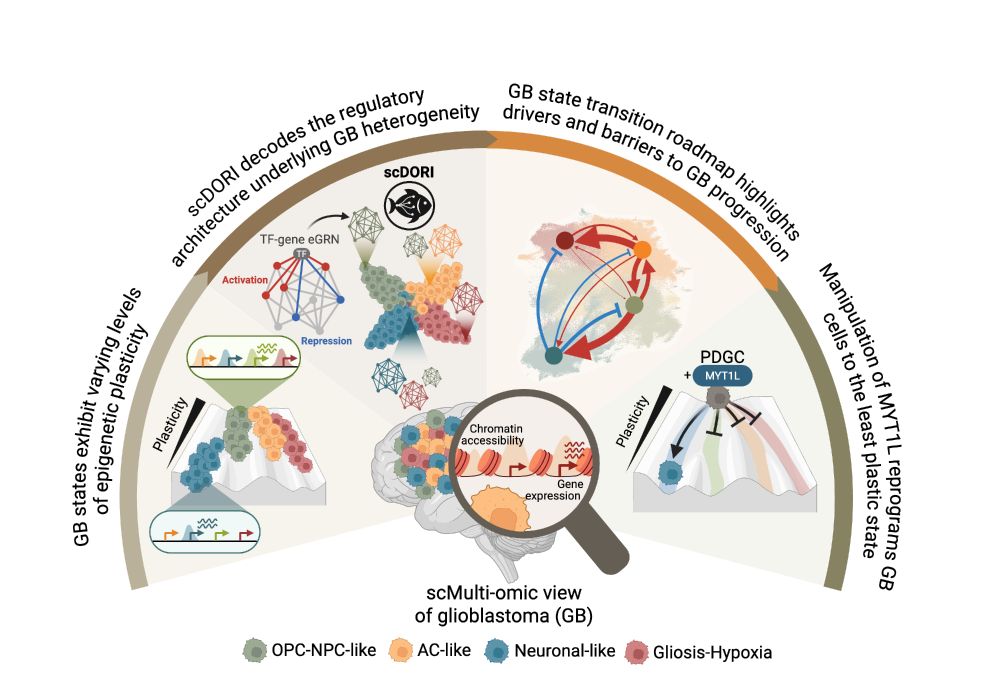
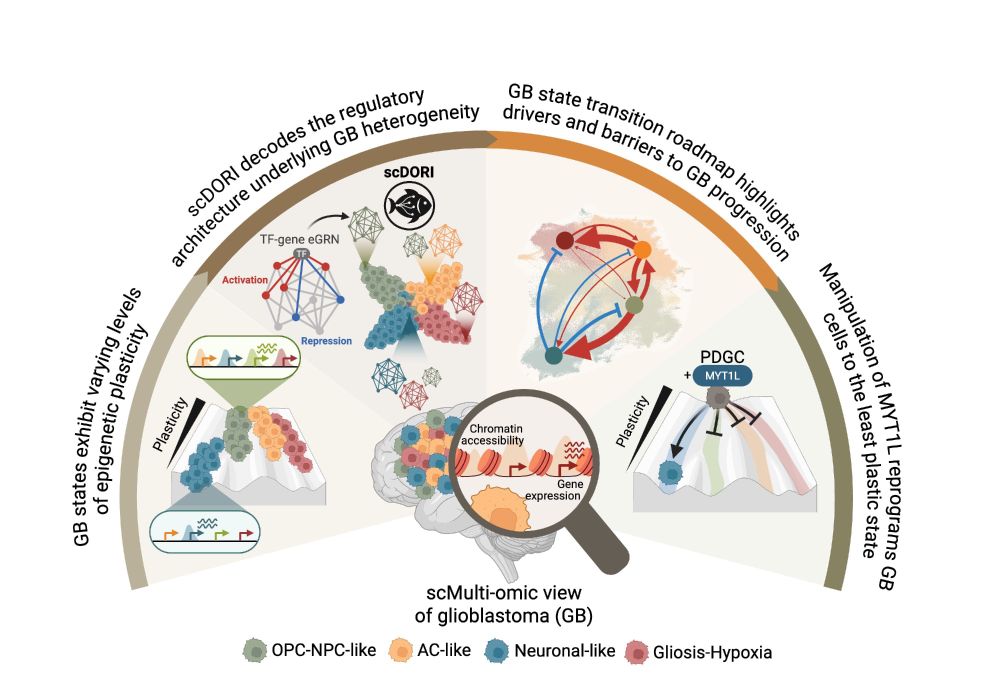
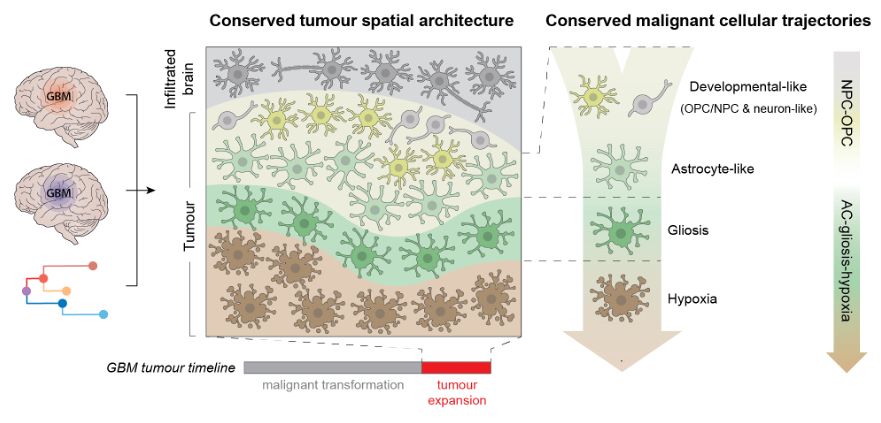
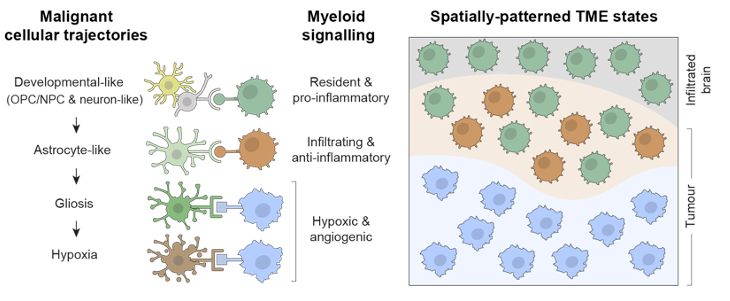
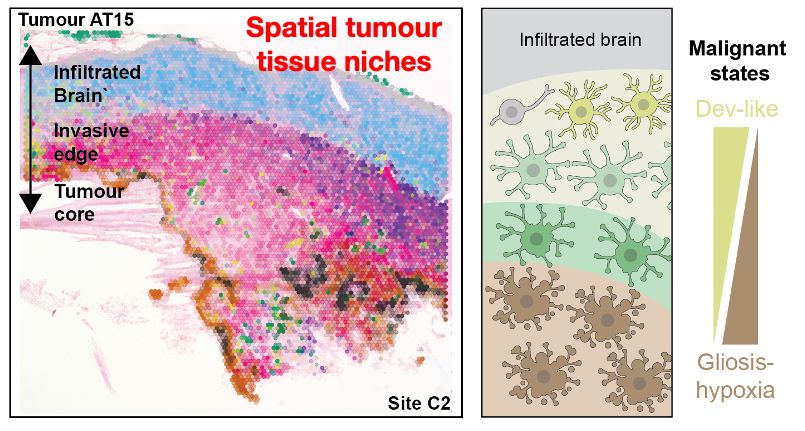
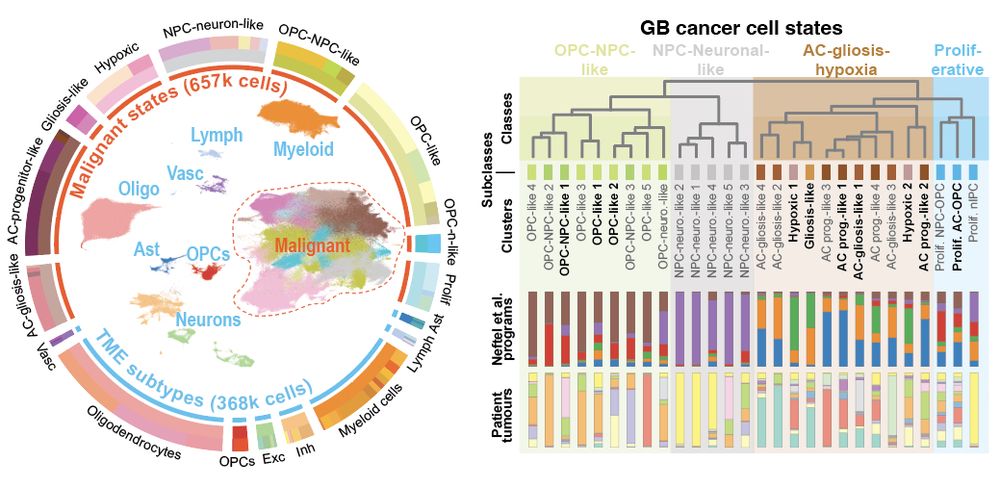
Decoding Plasticity Regulators and Transition Trajectories in Glioblastoma with Single-cell Multiomics
Glioblastoma (GB) is one of the most lethal human cancers, marked by profound intratumoral heterogeneity and near-universal treatment resistance. Cellular plasticity, the capacity of cancer cells to t...
www.biorxiv.org