Matthieu Schapira
@mattschap.bsky.social
890 followers
90 following
15 posts
Prof @ U. Toronto - PI @ SGC - CACHE challenges - Computational chemistry- Structural bioinformatics - Biophysics
Posts
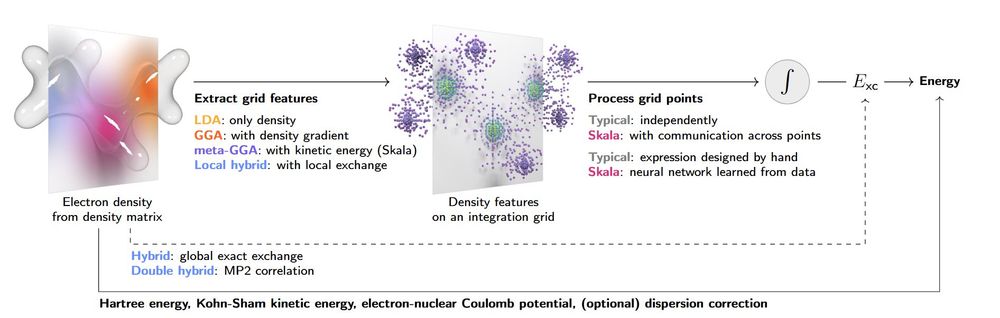
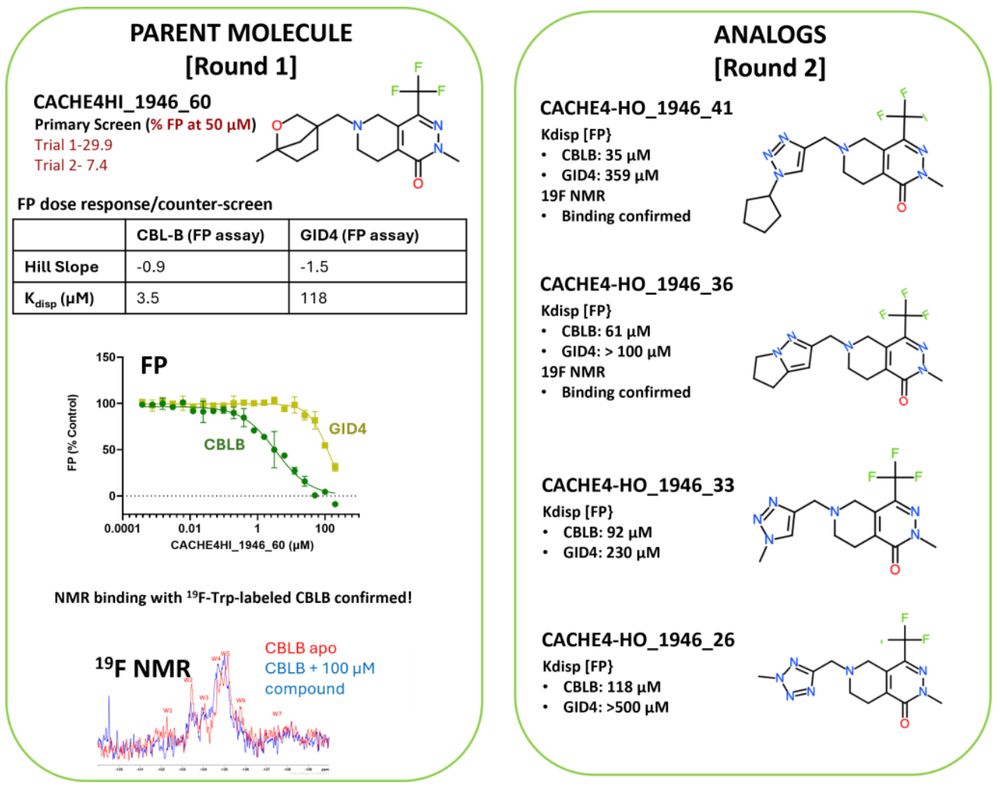
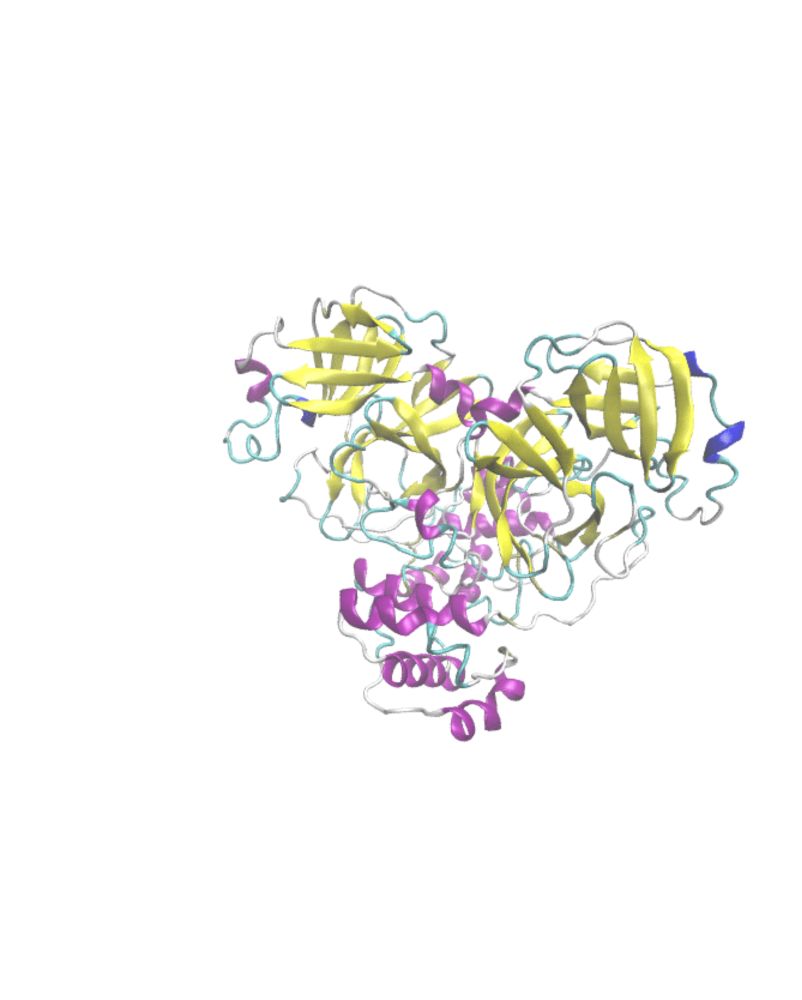
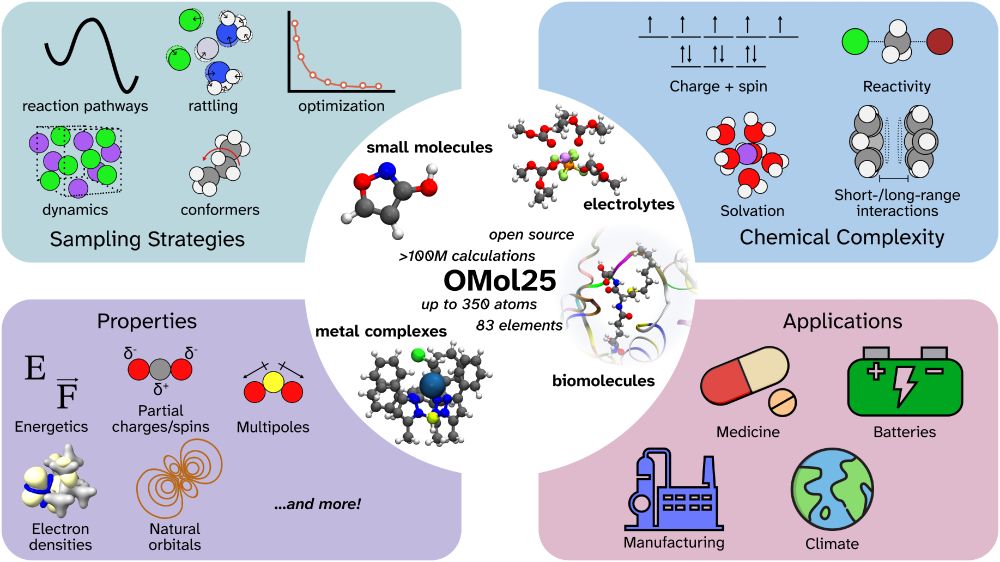
Media
Videos
Starter Packs
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Chris de Graaf
@cdg-gpcr.bsky.social
· Jul 2
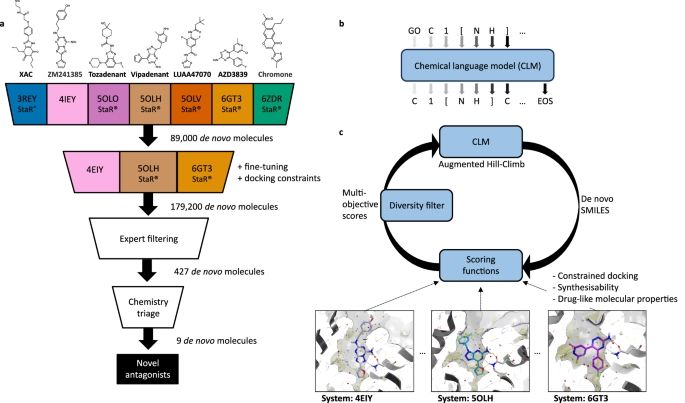
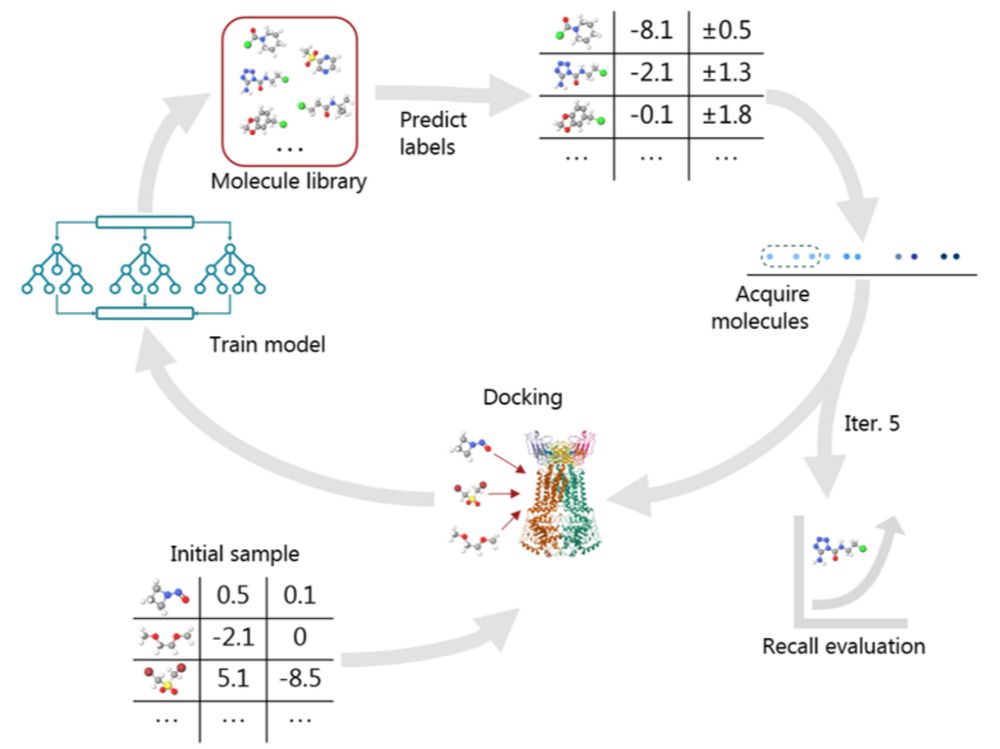
Identification of nanomolar adenosine A2A receptor ligands using reinforcement learning and structure-based drug design - Nature Communications
Here the authors combine a deep generative model with structure-based drug design and prospectively validate functionally active, nanomolar, A2A adenosine receptor ligands and solve their crystal stru...
www.nature.com
Matthieu Schapira
@mattschap.bsky.social
· Jun 20
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
Reposted by Matthieu Schapira
E. Richard Gold
@richardgold.bsky.social
· Apr 10

AI drug development’s data problem
The future of drug discovery may be artificial intelligence (AI), but its present is not. AI is in its infancy in the field. To help AI mature, developers need nonproprietary, open, large, high-qualit...
www.science.org
Reposted by Matthieu Schapira