Mike Clark
@michaelbclark.bsky.social
63 followers
48 following
10 posts
Genetics, transcriptomics, RNA and neuroscience.
Lab head at the University of Melbourne, Australia.
View own.
Posts
Media
Videos
Starter Packs
Reposted by Mike Clark
Reposted by Mike Clark
Reposted by Mike Clark
Mike Clark
@michaelbclark.bsky.social
· Aug 12
Mike Clark
@michaelbclark.bsky.social
· Aug 12
Reposted by Mike Clark
Science Magazine
@science.org
· Aug 10
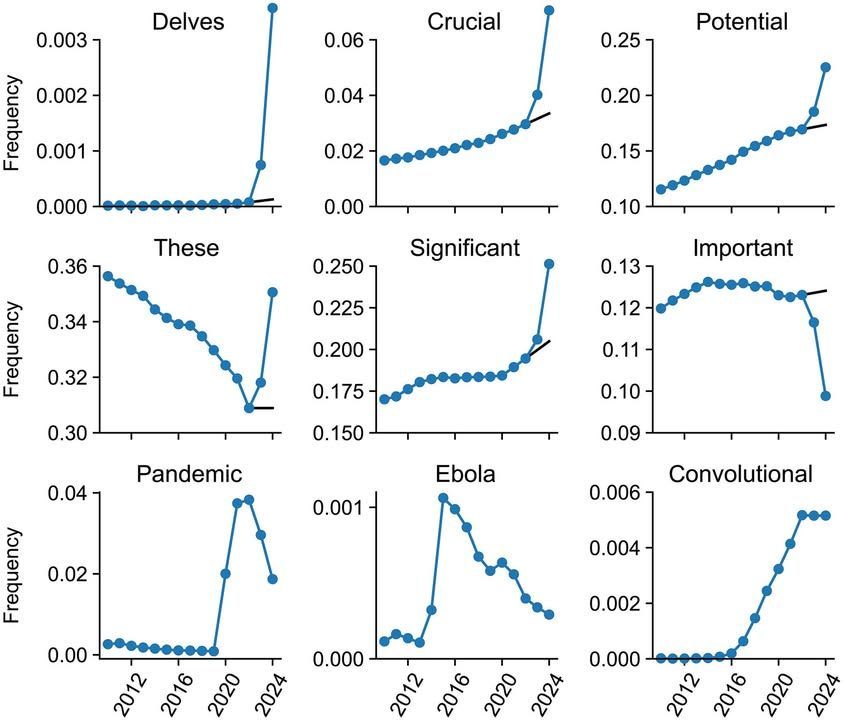
Delving into LLM-assisted writing in biomedical publications through excess vocabulary
Large language models (LLMs) like ChatGPT can generate and revise text with human-level performance. These models come with clear limitations, can produce inaccurate information, and reinforce existing biases.
scim.ag
Reposted by Mike Clark
Haley Lab
@labohaley.bsky.social
· Dec 21

A CRISPR/Cas9 screen reveals proteins at the endosome-Golgi interface that modulate cellular ASO activity
Anti-sense oligonucleotides (ASOs) are modified synthetic single-stranded molecules with enhanced stability, activity, and bioavailability. They associate with RNA through sequence complementarity and...
www.biorxiv.org








