Mitch Guttman
@mitchguttman.bsky.social
1.4K followers
110 following
15 posts
Molecular biologist interested in non-coding RNAs, nuclear organization, and gene regulation. Professor at Caltech
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Mitch Guttman
Mitch Guttman
@mitchguttman.bsky.social
· Jul 26
Mitch Guttman
@mitchguttman.bsky.social
· Nov 27
Mitch Guttman
@mitchguttman.bsky.social
· Nov 27
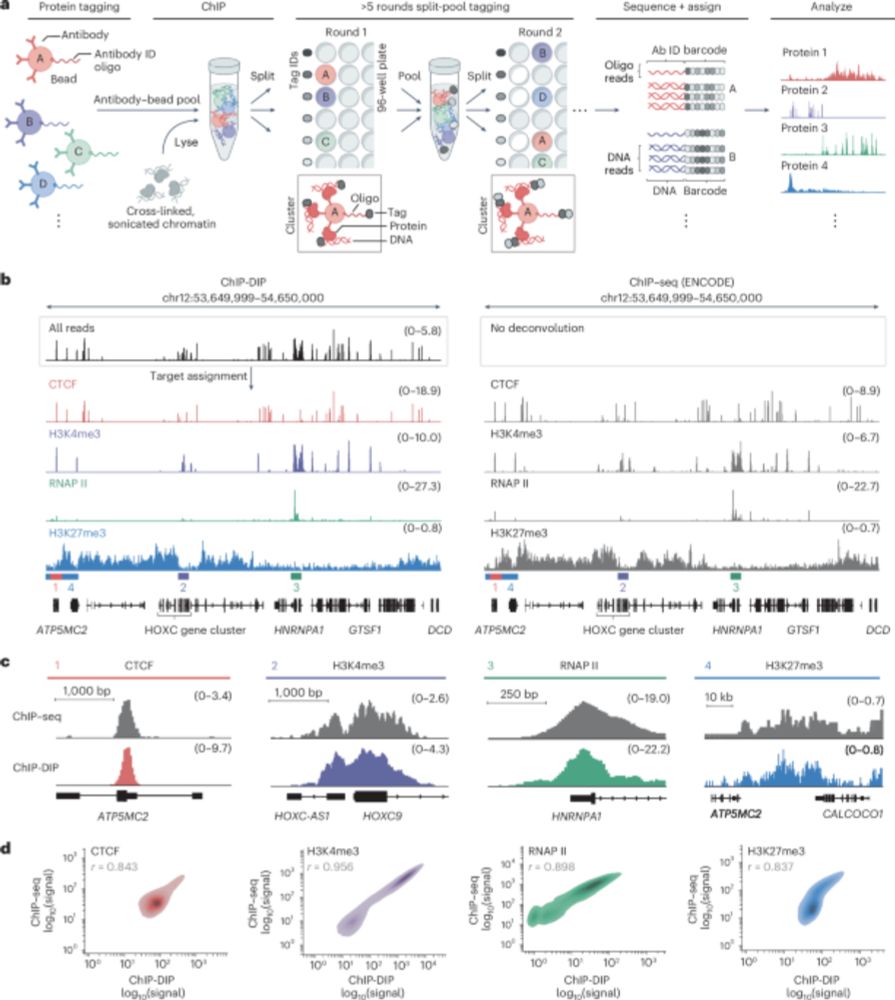
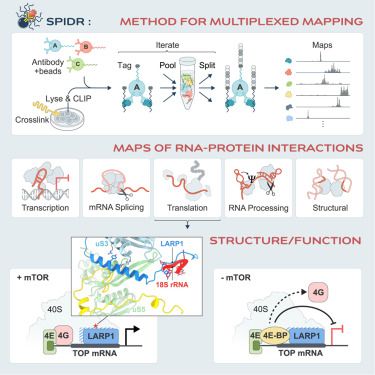
ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements - Nature Genetics
ChIP-DIP (ChIP done in parallel) is a highly multiplex assay for protein–DNA binding, scalable to hundreds of proteins including modified histones, chromatin regulators and transcription factors, offe...
www.nature.com
Reposted by Mitch Guttman
Mitch Guttman
@mitchguttman.bsky.social
· Nov 18

Failing to account for RNA quantity inflates background and leads to the misleading appearance that PRC2 and GFP bind to RNA in vivo
We recently published biochemical and quantitative evidence that challenges the widespread claims that PRC2 binds directly to many RNAs in vivo . A recent preprint performs a re-analysis of some of ou...
www.biorxiv.org
Reposted by Mitch Guttman