Paul Thomas
@pgtimmune.bsky.social
2.9K followers
610 following
64 posts
Department of Host-Microbe Interactions, St. Jude Children’s Research Hospital
Memphis, TN
TCRs, influenza virus, anti-tumor immunity, books, dogs, Venice
Posts
Media
Videos
Starter Packs
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14
Paul Thomas
@pgtimmune.bsky.social
· Jul 14

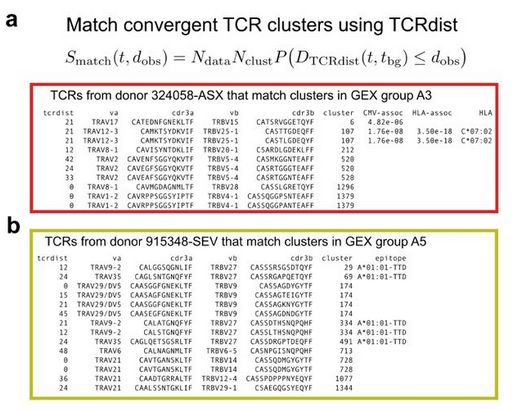
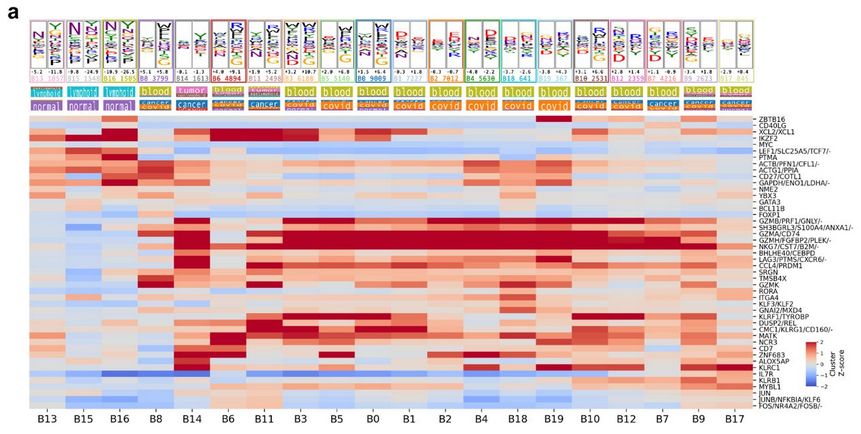
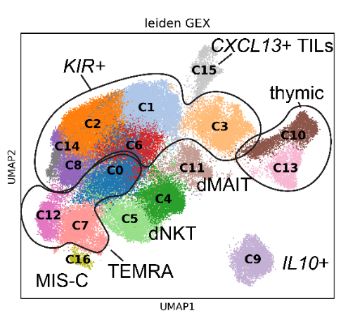
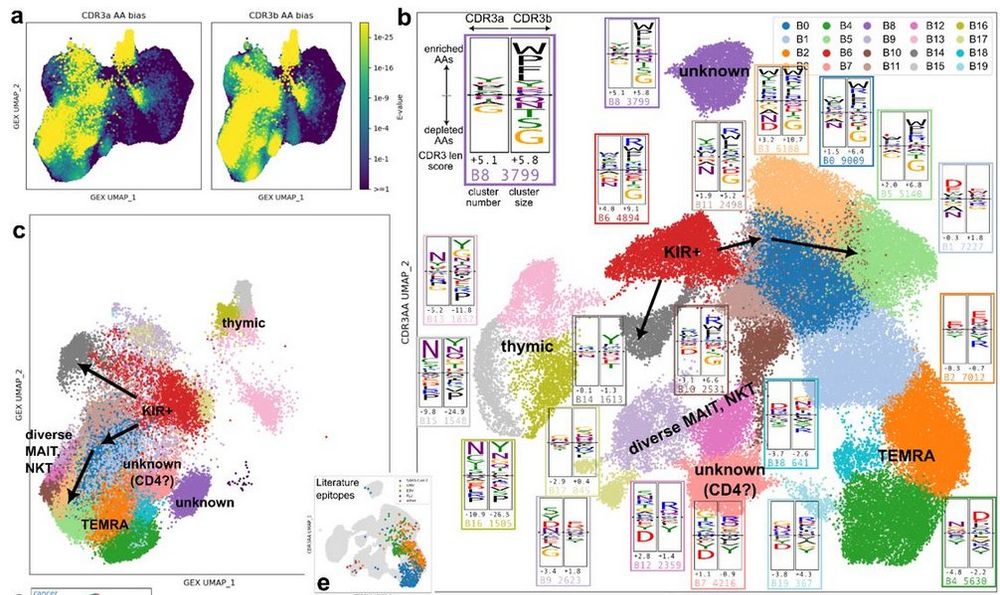
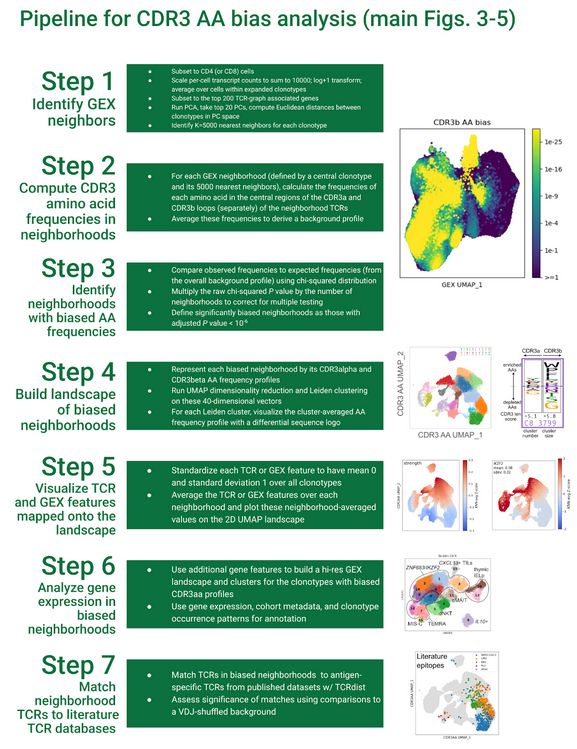
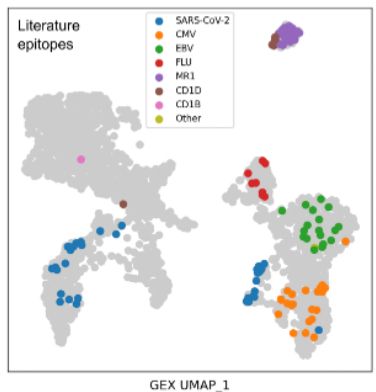
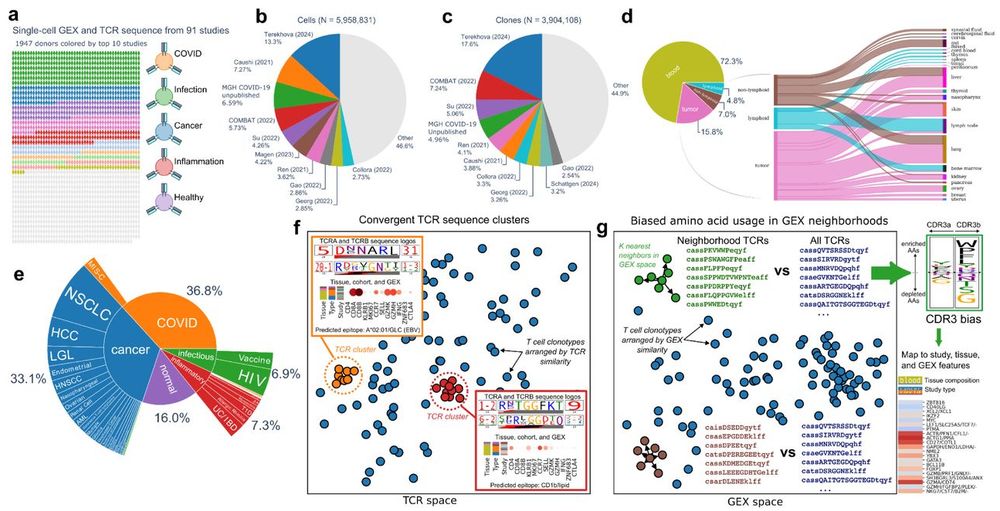
Diverse modes of T cell receptor sequence convergence define unique functional and cellular phenotypes
Single-cell techniques allow concurrent study of gene activity and T cell receptor (TCR) sequences, identifying connections between TCR structure and cell traits. Expanding on our CoNGA software, we p...
www.biorxiv.org
Reposted by Paul Thomas
David M. Perry
@lollardfish.bsky.social
· Apr 14
Paul Thomas
@pgtimmune.bsky.social
· Apr 4
Paul Thomas
@pgtimmune.bsky.social
· Mar 29
Reposted by Paul Thomas
Reposted by Paul Thomas
Stacey Ogden 🦔
@labogden.bsky.social
· Mar 28
Reposted by Paul Thomas
Reposted by Paul Thomas