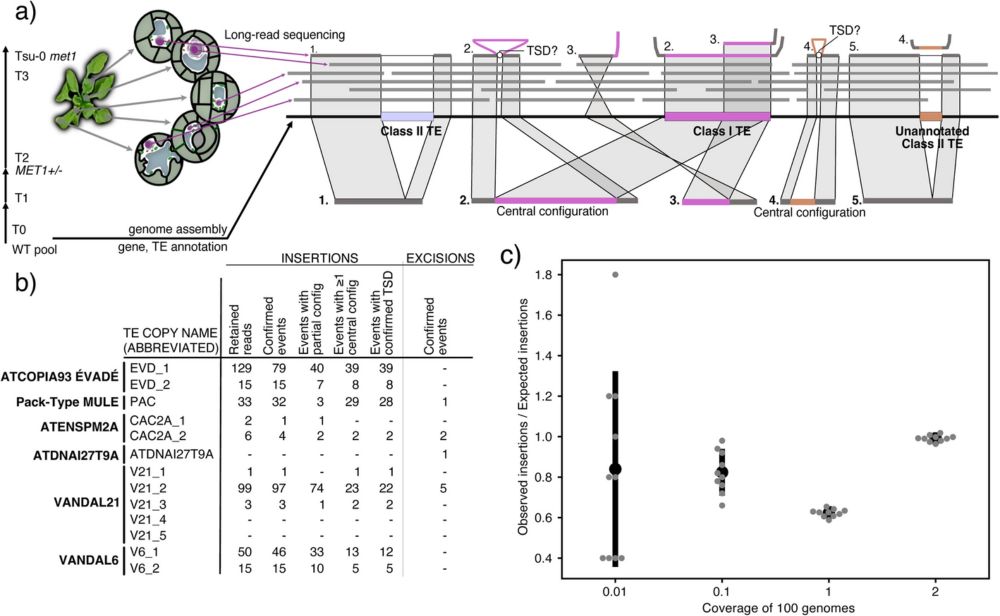
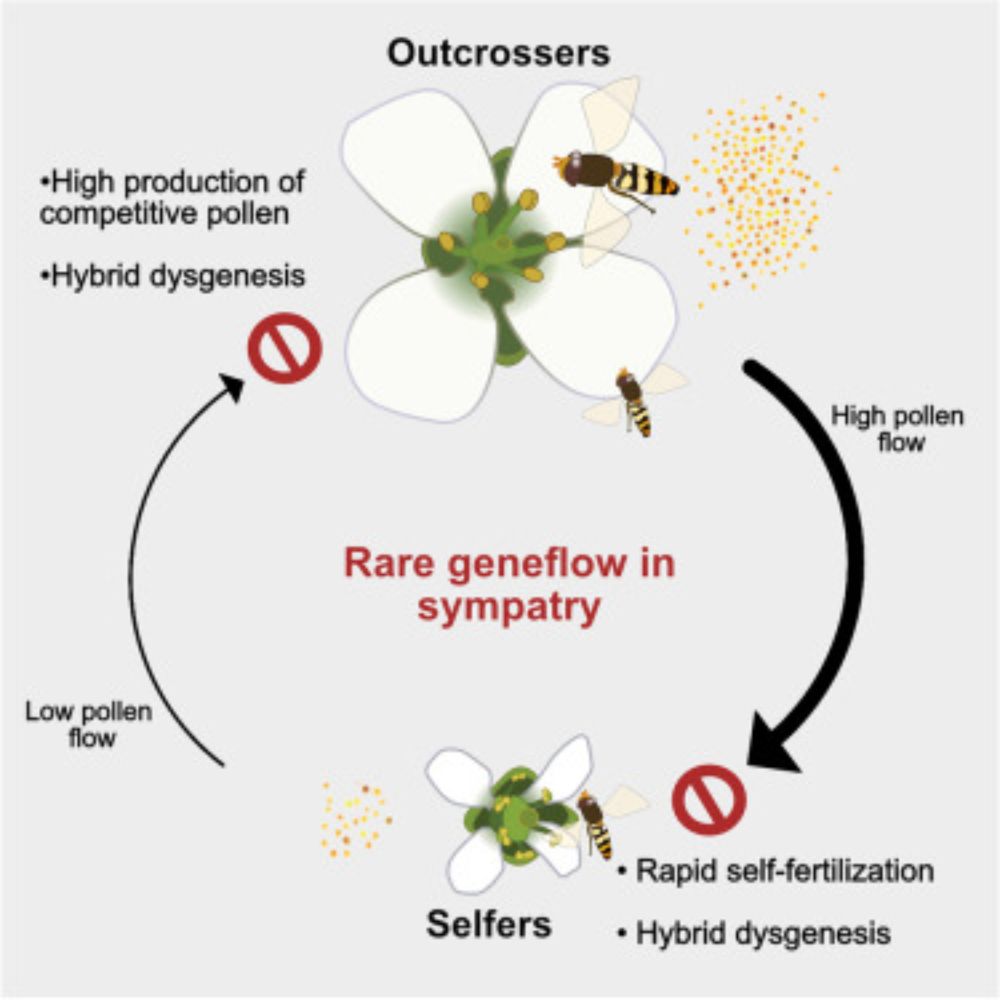
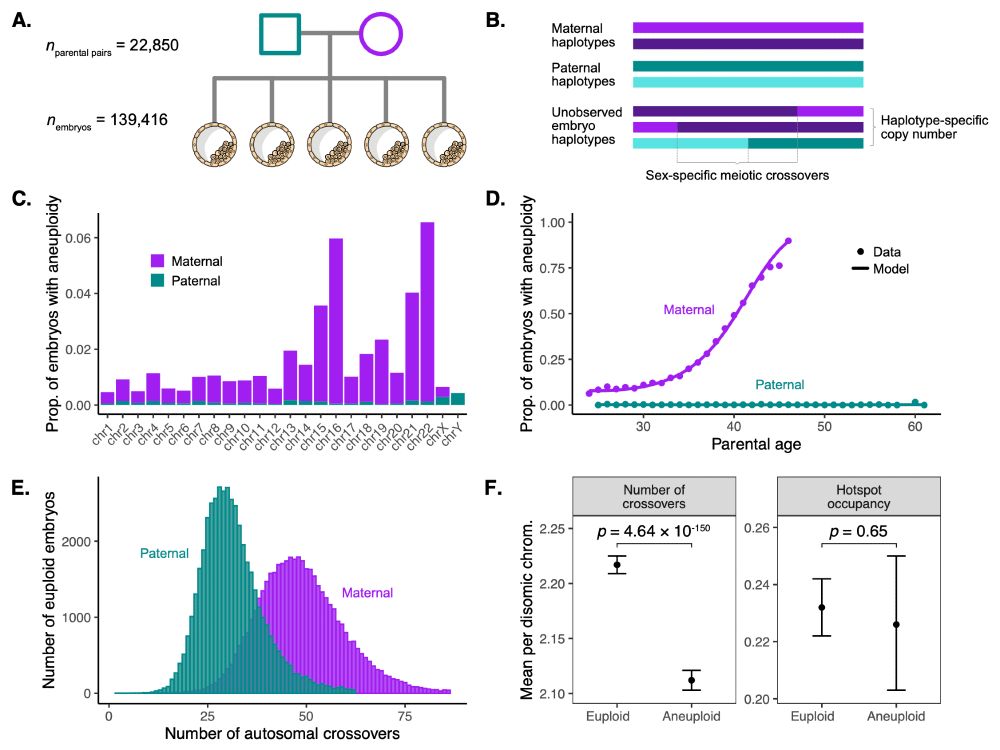
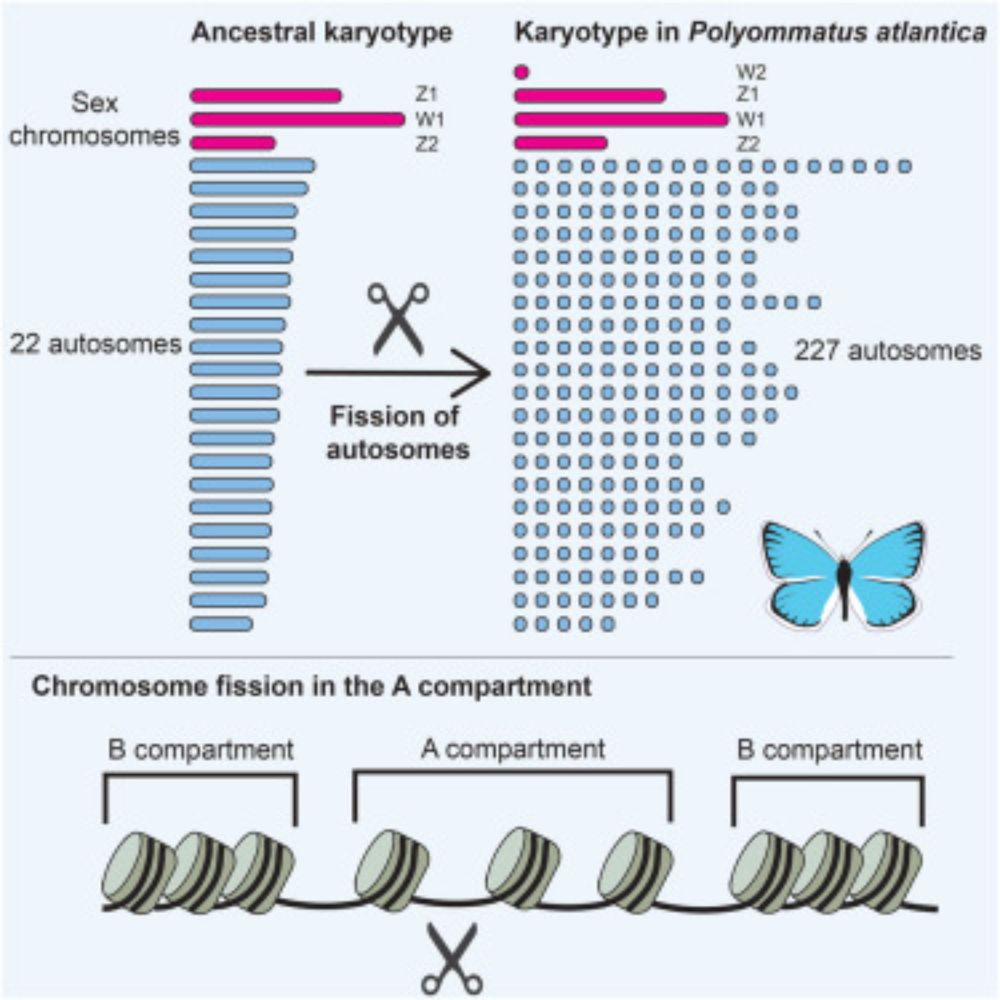
Piotr Ziolkowski
@piotraz.bsky.social
370 followers
860 following
19 posts
Geneticist interested in (meiotic) recombination and impact of chromatin structure on DNA functions
http://dgb.amu.edu.pl
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Piotr Ziolkowski
Reposted by Piotr Ziolkowski
Reposted by Piotr Ziolkowski
Reposted by Piotr Ziolkowski
Etienne Bucher 🌾🧬🇪🇺
@plantepigenetics.ch
· Aug 11

Plants repress ROS1 expression to attenuate heat-induced transposon burst - Nature Plants
Plants suppress the DNA demethylase ROS1 under heat stress to limit transposon activation, preventing genome instability. This conserved mechanism balances stress adaptation with genome protection.
www.nature.com
Reposted by Piotr Ziolkowski
Reposted by Piotr Ziolkowski
Reposted by Piotr Ziolkowski
Piotr Ziolkowski
@piotraz.bsky.social
· Sep 7
Reposted by Piotr Ziolkowski
Piotr Ziolkowski
@piotraz.bsky.social
· Sep 5
Piotr Ziolkowski
@piotraz.bsky.social
· Sep 2
Piotr Ziolkowski
@piotraz.bsky.social
· Sep 2

Enhancing local meiotic crossovers in Arabidopsis and maize through juxtaposition of heterozygous and homozygous regions - Nature Plants
This study discovered that locally confined DNA differences boost crossover rates in both Arabidopsis and maize, revealing a conserved mechanism that can accelerate plant breeding and trait introgress...
www.nature.com