Shinichi Nakagawa
@smoltblue.bsky.social
78 followers
12 following
10 posts
noncoding RNA
Developmental Biology
Molecular Biology
Hokkaido University
Posts
Media
Videos
Starter Packs
Pinned
Shinichi Nakagawa
@smoltblue.bsky.social
· Nov 15

Quick and affordable DNA cloning by reconstitution of Seamless Ligation Cloning Extract using defined factors
Molecular mechanisms underlying the widely accepted Seamless Ligation Cloning Extract method were elucidated. A novel protocol was developed for quick and affordable seamless DNA cloning. Commercial ....
onlinelibrary.wiley.com
Shinichi Nakagawa
@smoltblue.bsky.social
· Feb 20

Transposon–host arms race: a saga of genome evolution
Once considered ‘junk DNA,’ transposons or transposable elements (TEs) are now recognized
as key drivers of genome evolution, contributing to genetic diversity, gene regulation,
and species diversific...
www.cell.com
Reposted by Shinichi Nakagawa
Fyodor Urnov
@urnov.bsky.social
· Nov 29

Molecular and genetic characterization of sex-linked orange coat color in the domestic cat
The Sex-linked orange mutation in domestic cats causes variegated patches of reddish/yellow hair and is a defining signature of random X-inactivation in female tortoiseshell and calico cats. Unlike th...
www.biorxiv.org
Reposted by Shinichi Nakagawa
Reposted by Shinichi Nakagawa
Josh Mendell
@mendell-lab.bsky.social
· Nov 22
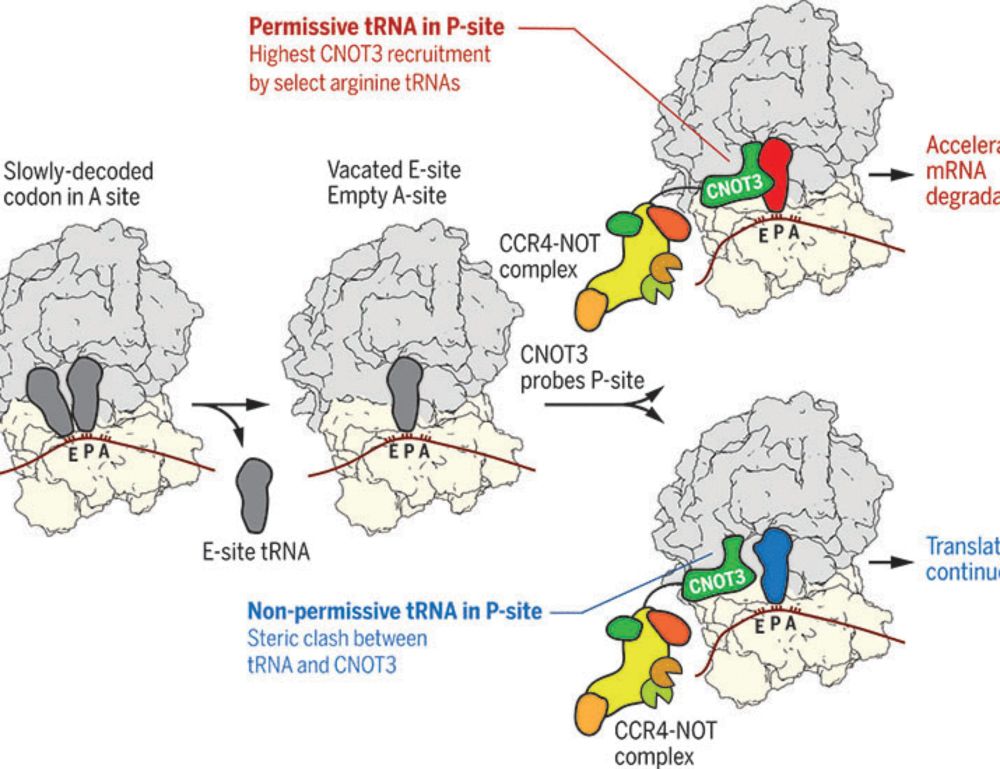
Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes
The CCR4-NOT complex is a major regulator of eukaryotic messenger RNA (mRNA) stability. Slow decoding during translation promotes association of CCR4-NOT with ribosomes, accelerating mRNA degradation....
www.science.org
Reposted by Shinichi Nakagawa
Shinichi Nakagawa
@smoltblue.bsky.social
· Nov 15
Shinichi Nakagawa
@smoltblue.bsky.social
· Nov 15
Shinichi Nakagawa
@smoltblue.bsky.social
· Nov 15

Quick and affordable DNA cloning by reconstitution of Seamless Ligation Cloning Extract using defined factors
Molecular mechanisms underlying the widely accepted Seamless Ligation Cloning Extract method were elucidated. A novel protocol was developed for quick and affordable seamless DNA cloning. Commercial ....
onlinelibrary.wiley.com