Soham Mukhopadhyay
@sohambio.bsky.social
140 followers
110 following
19 posts
Plant-Microbe Interaction | Postdoc @Edel's Lab @UniversiteLaval, Canada | Genomics | Aquascaping | Creator of biosearch.chat
Posts
Media
Videos
Starter Packs
Pinned
Soham Mukhopadhyay
@sohambio.bsky.social
· Feb 21
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Gang Yu
@gangyuplant.bsky.social
· Sep 1

Inactivation of β-1,3-glucan synthase-like 5 confers broad-spectrum resistance to Plasmodiophora brassicae pathotypes in cruciferous plants - Nature Genetics
This study implicates GSL5 inactivation in high, broad-spectrum resistance to the clubroot pathogen Plasmodiophora brassicae in Arabidopsis thaliana, Brassica napus, Brassica oleracea and Brassica rap...
www.nature.com
Reposted by Soham Mukhopadhyay
Soham Mukhopadhyay
@sohambio.bsky.social
· Aug 26
Soham Mukhopadhyay
@sohambio.bsky.social
· Aug 26
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Fantin Mesny
@mesny.bsky.social
· Aug 15

Plant-associated fungi co-opt ancient antimicrobials for host manipulation
Evolutionary histories of effector proteins secreted by fungal pathogens to mediate plant colonization remain largely elusive. While most functionally characterized effectors modulate plant immunity, ...
www.biorxiv.org
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Sorek Lab
@soreklab.bsky.social
· Jul 30
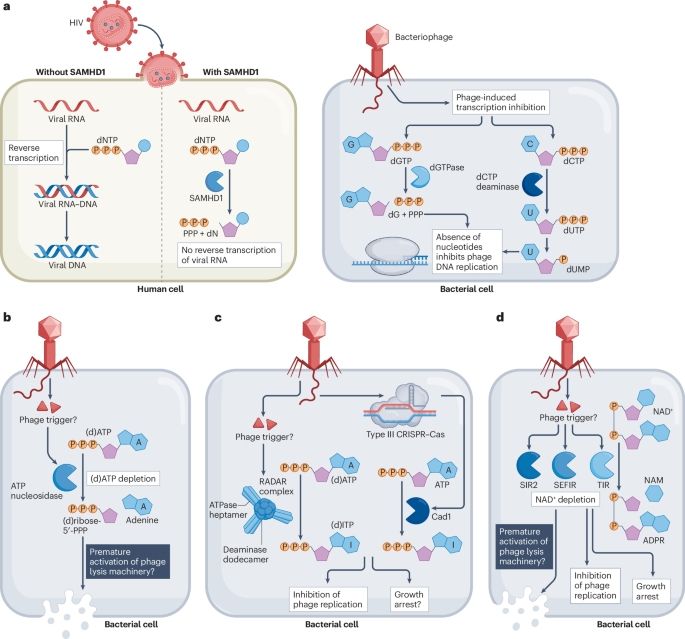
Manipulation of the nucleotide pool in human, bacterial and plant immunity - Nature Reviews Immunology
Modification of the nucleotide pool is emerging as key to innate immunity in animals, plants and bacteria. This Review explains how immune pathways conserved from bacteria to humans manipulate the nuc...
www.nature.com
Reposted by Soham Mukhopadhyay
Sorek Lab
@soreklab.bsky.social
· Jul 25
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Tatsuya Nobori
@tatsuyanobori.bsky.social
· Jul 16
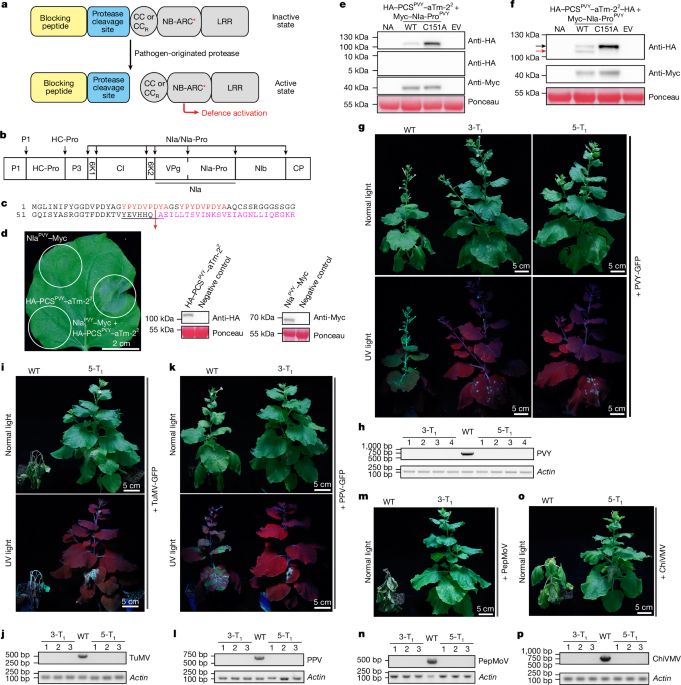
Remodelling autoactive NLRs for broad-spectrum immunity in plants - Nature
Cleavage by pathogen-derived proteases of an engineered chimeric protein activates its plant immune receptor component, enabling broad-spectrum resistance to pathogens in plants.
www.nature.com
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Yu Sugihara
@yusugihara.bsky.social
· Jul 16
Reposted by Soham Mukhopadhyay
Reposted by Soham Mukhopadhyay
Sebastian Schornack
@dromius.bsky.social
· Jul 13

The N-terminal domains of NLR immune receptors exhibit structural and functional similarities across divergent plant lineages
Plant immunity mediated by a major class of immune receptors is conserved among flowering and nonflowering plants, and these receptors have domains with si
academic.oup.com