Sukrit Singh
@sukritsingh92.bsky.social
3K followers
1.1K following
42 posts
Biophysicist|Damon Runyon QB & NCI Early Stage K99/R00 Fellow at MSKCC w/ @jchodera.bsky.social | @foldingathome.org | PhD with @drgregbowman.bsky.social | Variant Effects, Protein dynamics, Dogs, Games 🇮🇳🇸🇬🇺🇸
My website: https://sukritsingh.github.io
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Sukrit Singh
@sukritsingh92.bsky.social
· Jun 28
Reposted by Sukrit Singh
Vincent Voelz
@voelzlab.bsky.social
· Jun 23
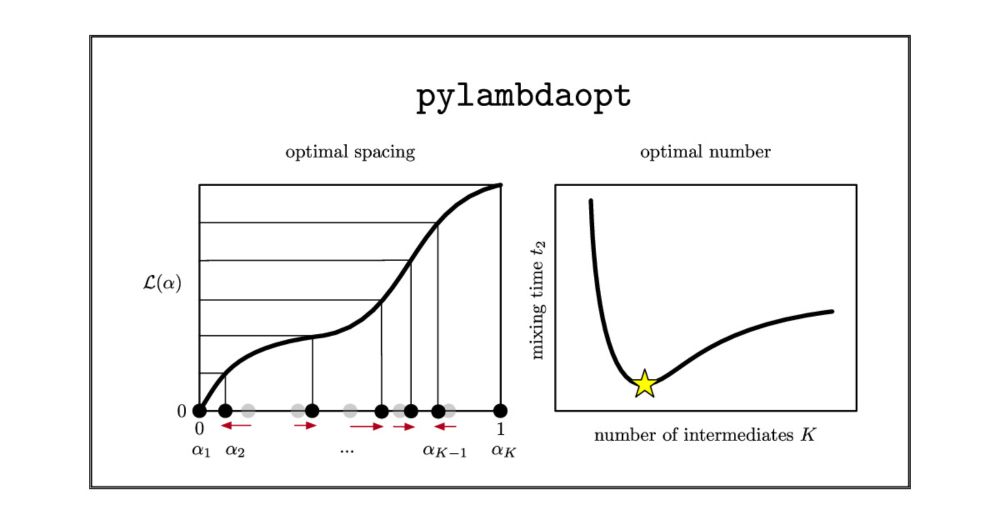
Simple Method to Optimize the Spacing and Number of Alchemical Intermediates in Expanded Ensemble Free Energy Calculations
Alchemical free energy calculations are essential to modern structure-based drug design. Such calculations are usually performed at a series of discrete intermediates along a nonphysical thermodynamic...
doi.org
Reposted by Sukrit Singh
Reposted by Sukrit Singh
John Chodera
@jchodera.bsky.social
· Mar 25
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Reposted by Sukrit Singh
Mark Polk
@markpolk.io
· Feb 16