Tilmann Weber
@tilmweber.bsky.social
900 followers
1.4K following
21 posts
Professor at DTU NNF Center for Biosustainability with interest in bioactive compounds, comp. biol., WGS and much more; hobby photographer. Views are my own.
Posts
Media
Videos
Starter Packs
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Kai Blin
@kblin.bsky.social
· May 26
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Mingxun Wang
@mingxunwang.bsky.social
· May 12
Reposted by Tilmann Weber
Reposted by Tilmann Weber
Tilmann Weber
@tilmweber.bsky.social
· Apr 28
Tilmann Weber
@tilmweber.bsky.social
· Apr 28
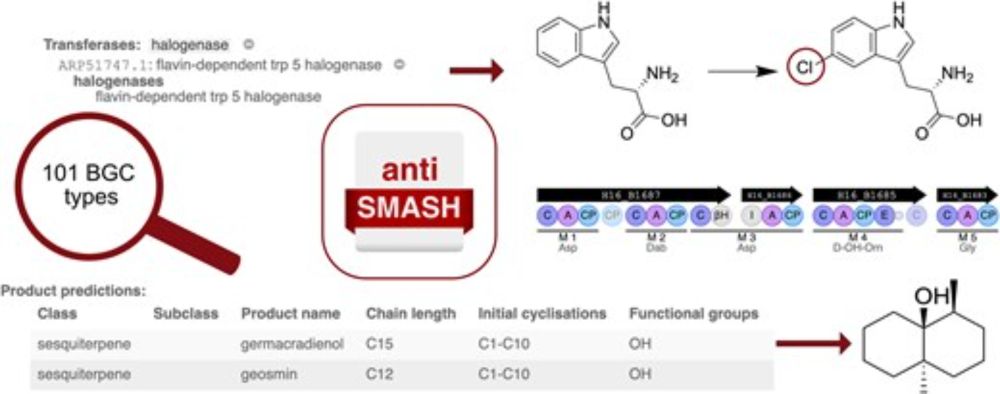
antiSMASH 8.0: extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation
Abstract. Microorganisms synthesize small bioactive compounds through their secondary or specialized metabolism. Those compounds play an important role in
academic.oup.com
Reposted by Tilmann Weber
Tilmann Weber
@tilmweber.bsky.social
· Apr 5
Tilmann Weber
@tilmweber.bsky.social
· Apr 5

ActinoMation: A literate programming approach for medium-throughput robotic conjugation of Streptomyces spp
The genus Streptomyces are valuable producers of antibiotics and other pharmaceutically important bioactive compounds. Advances in molecular engineeri…
www.sciencedirect.com
Reposted by Tilmann Weber
Reposted by Tilmann Weber














