Julia M. Brown
@viraljules.bsky.social
260 followers
480 following
7 posts
(mostly) marine microbial and viral ecology via all the 'omics
Mainer from away
check out my lab webpage at juliambrown.github.io
Posts
Media
Videos
Starter Packs
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Sean Gibbons 🦠💩
@gibbological.bsky.social
· Jul 31
Metagenomic estimation of absolute bacterial biomass in the mammalian gut through host-derived read normalization | mSystems
In this study, we asked whether normalization by host reads alone was sufficient to
estimate absolute bacterial biomass directly from stool metagenomic data, without
the need for synthetic spike-ins, ...
journals.asm.org
Reposted by Julia M. Brown
Joe Zackular
@joeyzacks.bsky.social
· Aug 20
Reposted by Julia M. Brown
George Bouras
@gbouras13.bsky.social
· Aug 8

Protein Structure Informed Bacteriophage Genome Annotation with Phold
Bacteriophage (phage) genome annotation is essential for understanding their functional potential and suitability for use as therapeutic agents. Here we introduce Phold, an annotation framework utilis...
www.biorxiv.org
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Julia M. Brown
@viraljules.bsky.social
· Jul 17
Reposted by Julia M. Brown
Emil Ruff
@emilruff.bsky.social
· Jun 2

Simons Graduate Fellowships in Ecology and Evolution
The purpose of these awards is to provide support for students entering U.S.-based Ph.D. programs with a plan to perform research in ecology and evolution. While we will consider all projects in ecolo...
www.simonsfoundation.org
Reposted by Julia M. Brown
Sean Eddy
@cryptogenomicon.bsky.social
· May 22
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Carl Zimmer
@carlzimmer.com
· May 1
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Cameron Thrash
@jcamthrash.bsky.social
· Jan 28
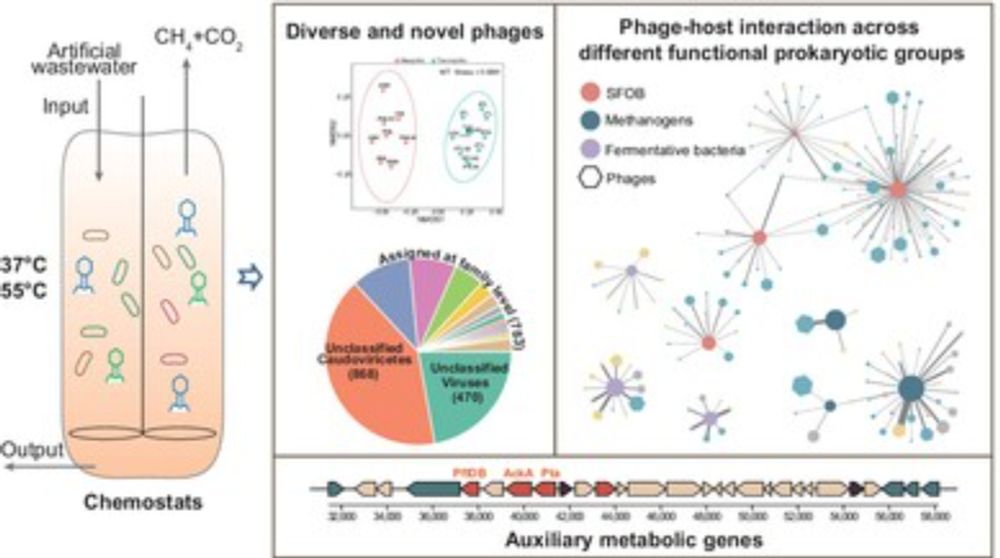
Characteristics of Phages and Their Interactions With Hosts in Anaerobic Reactors
Diverse and novel phages were recovered from anaerobic fatty acid–fed reactors. Phages linked to hosts in all three functional groups primarily consisted of generalists and temperate species, especia...
enviromicro-journals.onlinelibrary.wiley.com
Reposted by Julia M. Brown
Reposted by Julia M. Brown
Phage papers
@phagepapers.bsky.social
· Dec 1

Phage-induced disturbance of a marine sponge microbiome - PubMed
Our findings suggest that sponge microbiome diversity and, by extension, its resilience depend on the maintenance of resident bacterial community members, irrespective of their abundance. Phage-induced disturbances can significantly alter community structure by promoting the growth of opportunistic …
pubmed.ncbi.nlm.nih.gov
Reposted by Julia M. Brown
Rob Lanfear
@roblanfear.bsky.social
· Nov 26
Reposted by Julia M. Brown