Yousuf A. Khan
@yousufakhan.bsky.social
1.4K followers
250 following
40 posts
Group Leader @Stanford. RNA focused ML/AI, cryoEM/ET, and biophysics. Formerly:DeepMind AlphaFold,EvoscaleAI, Churchill Scholar@Cambridge_Uni & seen on Netflix
Posts
Media
Videos
Starter Packs
Pinned
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan (YAK) Lab - RNA Research at Stanford University
Yousuf A. Khan (YAK) Lab - Studying RNA folding, interactions, and cellular effects using Molecular and Cellular Physiology, Structural Biology, and Machine Learning at Stanford University
www.yousufakhan.com
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan
@yousufakhan.bsky.social
· Aug 27
Yousuf A. Khan (YAK) Lab - RNA Research at Stanford University
Yousuf A. Khan (YAK) Lab - Studying RNA folding, interactions, and cellular effects using Molecular and Cellular Physiology, Structural Biology, and Machine Learning at Stanford University
www.yousufakhan.com
Reposted by Yousuf A. Khan
Yousuf A. Khan
@yousufakhan.bsky.social
· Jul 29
Reposted by Yousuf A. Khan
Alex Rubinsteyn
@alexr.bsky.social
· Jul 23
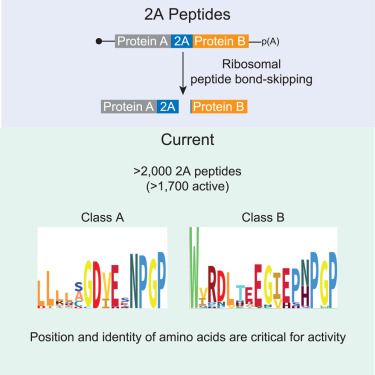
Systematic identification and characterization of eukaryotic and viral 2A peptide-bond-skipping sequences
Rao et al. identified thousands of previously unknown 2A peptides across both viruses
and eukaryotes using an HMMER analysis. The authors further identified a unique class
of 2A peptides, class B, who...
www.cell.com
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Arne Elofsson
@bioinfo.se
· May 5
Limits of deep-learning-based RNA prediction methods
Motivation: In recent years, tremendous advances have been made in predicting protein structures and protein-protein interactions. However, progress in predicting the structure of RNA, either alone or...
www.biorxiv.org
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan
Reposted by Yousuf A. Khan