Pedro Dorado-Morales
@doradomoralesp.bsky.social
180 followers
170 following
10 posts
MSCA fellow at PGB Unit (Institut Pasteur). Interested in bacterial genetics, bacterial signalling, mobile genetic elements and synthetic biology
Posts
Media
Videos
Starter Packs
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
Julie Le Bris
@julielebris.bsky.social
· Sep 10

Serotype swapping in Klebsiella spp. by plug-and-play
Understanding how complex, multi-gene systems evolve and function across genetic backgrounds is a central question in molecular evolution. While such systems often impose costs through epistatic inter...
www.biorxiv.org
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
David Sünderhauf
@davvi36.bsky.social
· Sep 2
Developing CRISPR-Cas antimicrobials to tackle antibiotic resistance spread in Klebsiella pneumoniae - GW4 BioMed MRC DTP
Project Code MRCIIAR26Ex van Houte Project Type Wet lab Research Theme Infection, Immunity, Antimicrobial Resistance and Repair Project Summary Download Summary Antimicrobial resistance (AMR) poses a ...
gw4biomed.ac.uk
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
Sarah Bigot
@sbigot.bsky.social
· Aug 20

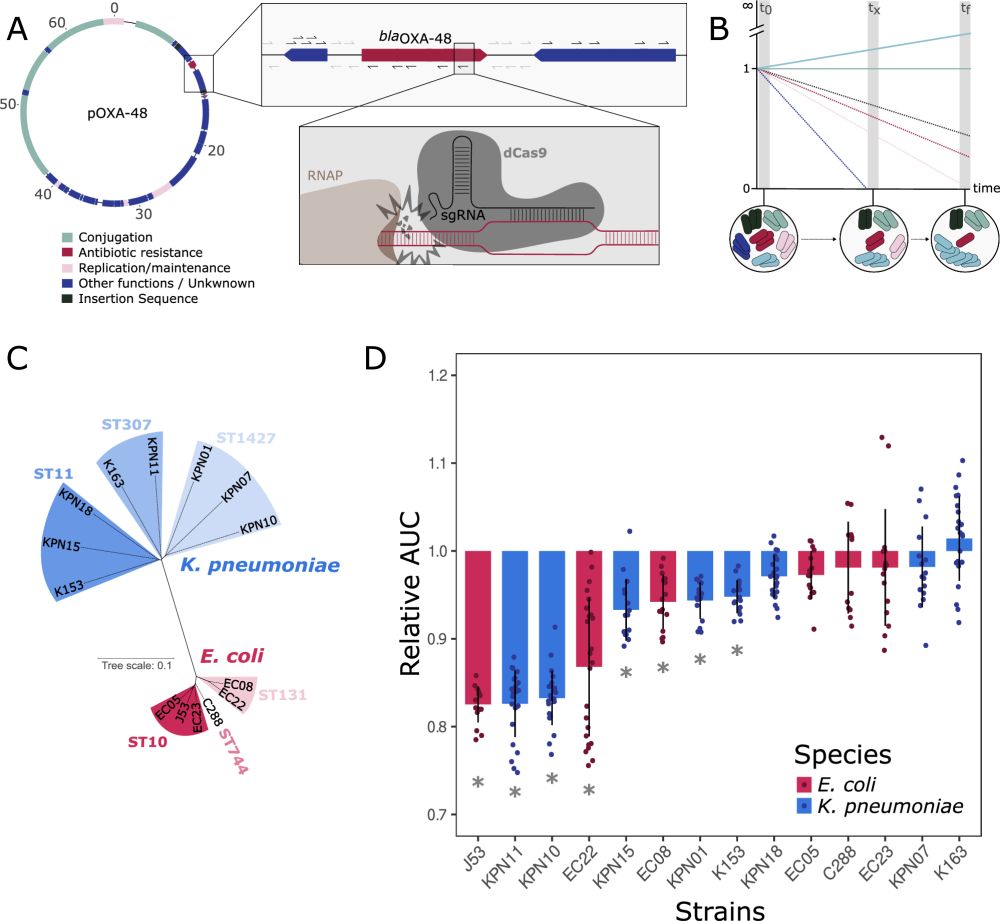
Genetic determinants of pOXA-48 plasmid maintenance and propagation in Escherichia coli - Nature Communications
pOXA-48 plasmids have emerged as key vectors of carbapenem resistance within Enterobacteriaceae. In this study, the authors use a transposon sequencing (Tn-seq) approach to identify genetic determinan...
www.nature.com
Reposted by Pedro Dorado-Morales
Didier Mazel
@amazeld.bsky.social
· Aug 20

Taming wild replicons: evolution and domestication of large extrachromosomal replicons
Bacterial genomes often contain extrachromosomal replicons (ERs), ranging from small, mobile plasmids to large, stably inherited elements, such as meg…
www.sciencedirect.com
Reposted by Pedro Dorado-Morales
Alvaro San Millan
@sanmillan.bsky.social
· Aug 13
Reposted by Pedro Dorado-Morales
Craig MacLean
@craigmaclean.bsky.social
· Jul 29

Plasmids link antibiotic resistance genes and phage defense systems in E. coli
Phage therapy has been proposed as an alternative to antibiotics to treat resistant infections. However, we have a limited understanding of how antibiotic resistance genes (ARGs) associate with bacter...
www.biorxiv.org
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales
Eugen Pfeifer
@eugenpfeifer.bsky.social
· Jul 28
Reposted by Pedro Dorado-Morales
Reposted by Pedro Dorado-Morales