


pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
#secmet #lassopeptides #syntheticbiology

#secmet #lassopeptides #syntheticbiology
pubs.acs.org/doi/10.1021/...
#secmet

pubs.acs.org/doi/10.1021/...
#secmet
pubs.acs.org/doi/10.1021/...

You like potato and I like potato
You like tomato and I like tomato
Potato, potahto, tomato, tomahto
Let's call the whole thing off
🧪
www.nytimes.com/2025/07/31/s...

You like potato and I like potato
You like tomato and I like tomato
Potato, potahto, tomato, tomahto
Let's call the whole thing off
🧪
@vanderbilt.edu @vubasicsciences.bsky.social

@vanderbilt.edu @vubasicsciences.bsky.social

pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
We enjoy knowing if metabolite is active before isolating it, and so should you! #natprod

We enjoy knowing if metabolite is active before isolating it, and so should you! #natprod
What happens behind the scenes with NIH fund transfers

What happens behind the scenes with NIH fund transfers




@standupforscience.bsky.social #standupforscience2025




@standupforscience.bsky.social #standupforscience2025
podcasts.apple.com/us/podcast/q...

podcasts.apple.com/us/podcast/q...
journals.asm.org/doi/10.1128/...
#secmet #MEvoSky

journals.asm.org/doi/10.1128/...
#secmet #MEvoSky
If this affects you call your reps & senators, talk to local news, make your voice heard 📢 👩🔬
www.youtube.com/watch?v=Cjdb...

www.youtube.com/watch?v=Cjdb...
pubs.acs.org/doi/10.1021/...
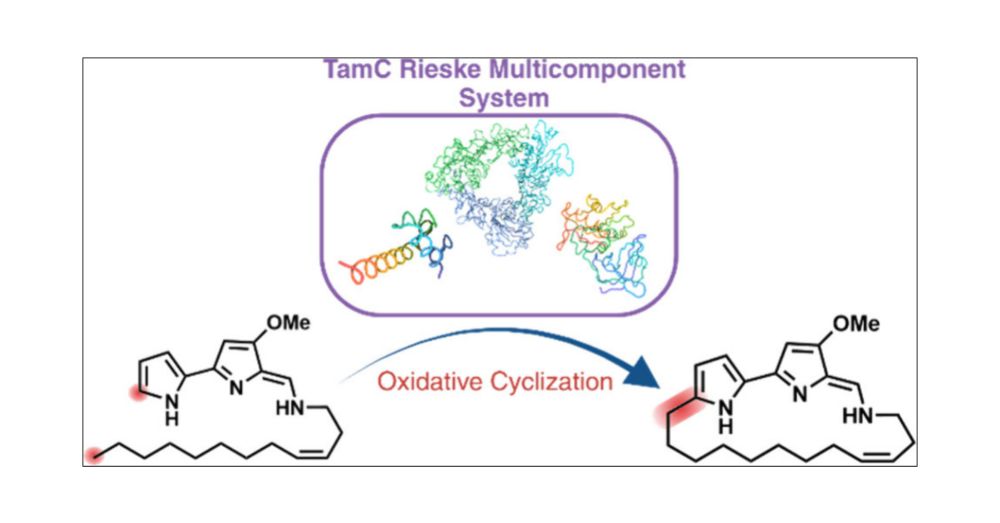
pubs.acs.org/doi/10.1021/...
Many thanks to the wonderful collaborators!
www.nature.com/articles/s41...

Many thanks to the wonderful collaborators!
www.nature.com/articles/s41...


