
Founding ingeniebio.com
ORCID 0000-0003-224
ow.ly/NJzp50WCc00 #NSF #AIwire

ow.ly/NJzp50WCc00 #NSF #AIwire
Head of Signal, Meredith Whittaker, on so-called "agentic AI" and the difference between how it's described in the marketing and what access and control it would actually require to work as advertised.
Head of Signal, Meredith Whittaker, on so-called "agentic AI" and the difference between how it's described in the marketing and what access and control it would actually require to work as advertised.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.


Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...

Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
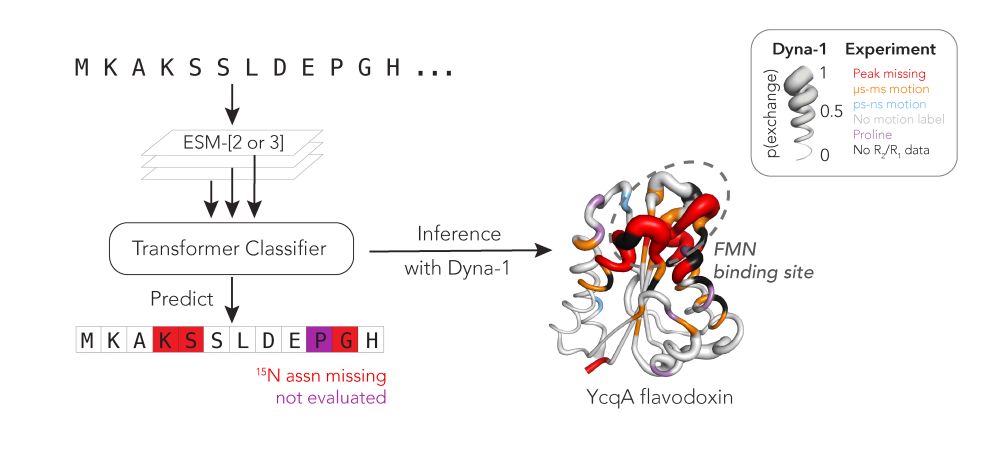
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
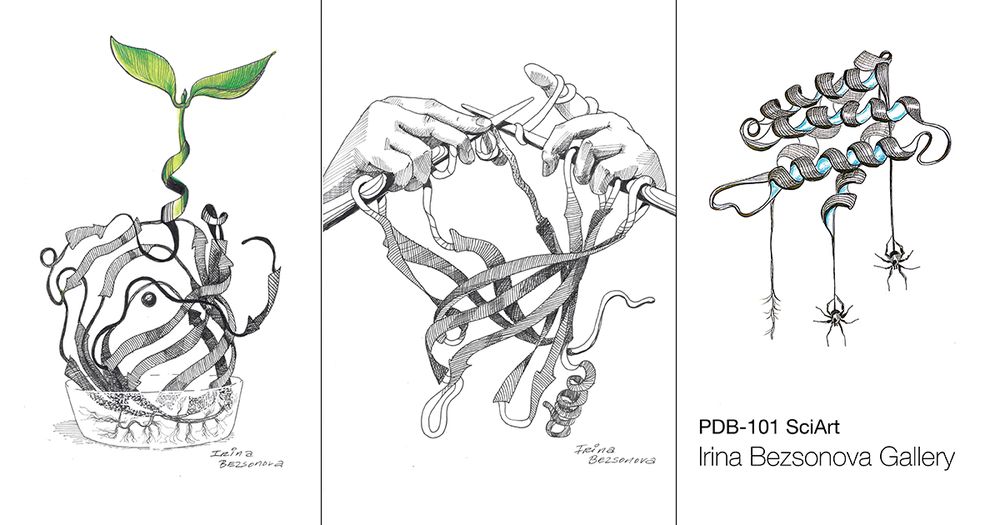
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
Jevin West (@jevinwest.bsky.social) and I have spent the last eight months developing the course on large language models (LLMs) that we think every college freshman needs to take.
thebullshitmachines.com

Jevin West (@jevinwest.bsky.social) and I have spent the last eight months developing the course on large language models (LLMs) that we think every college freshman needs to take.
thebullshitmachines.com
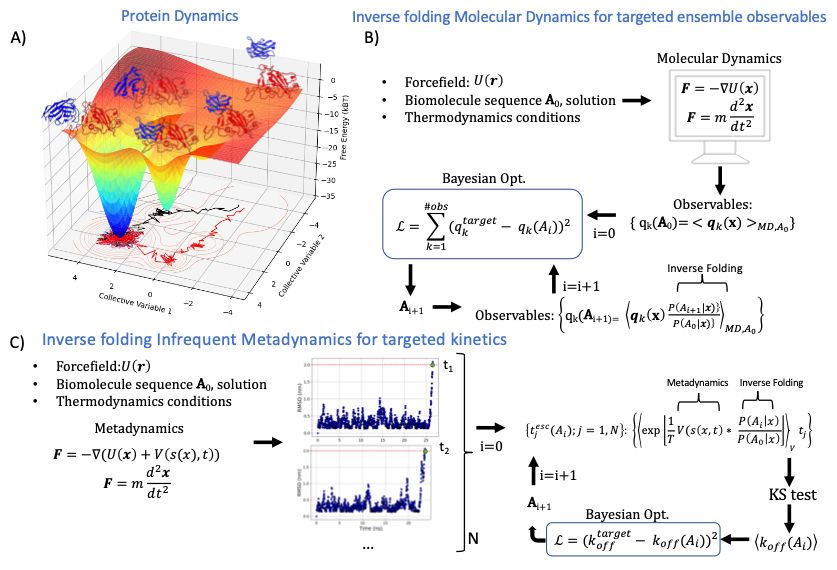
doi.org/10.1016/j.sb...
Brilliant and thoughtful piece.
doi.org/10.1016/j.sb...
Brilliant and thoughtful piece.
youtu.be/P_fHJIYENdI?...
youtu.be/P_fHJIYENdI?...
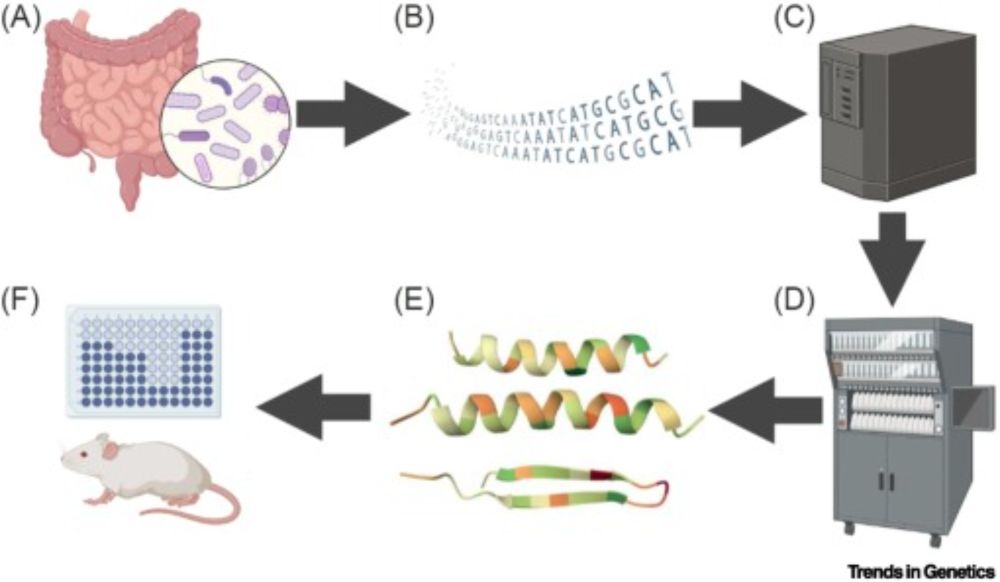
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
We introduce Venomics AI.
www.biorxiv.org/content/10.1...

We introduce Venomics AI.
www.biorxiv.org/content/10.1...
Just how many types of antibiotics were discovered during that time?
So, I visualized it myself!

Just how many types of antibiotics were discovered during that time?
So, I visualized it myself!





www.nature.com/articles/s41...
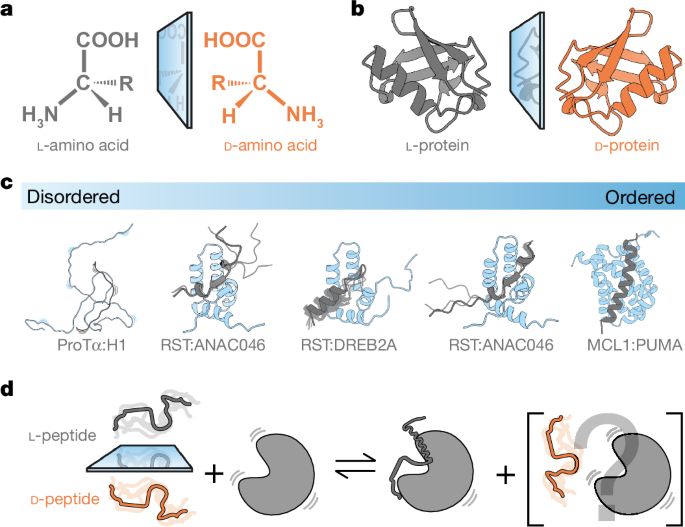

The programme is run jointly by the Universities of Bristol and Oxford.
Apply to Bristol (by 13 Jan 2025): bristol.ac.uk/study/postgr...
Apply to Oxford (by 8 Jan 2025): ox.ac.uk/admissions/g...

The programme is run jointly by the Universities of Bristol and Oxford.
Apply to Bristol (by 13 Jan 2025): bristol.ac.uk/study/postgr...
Apply to Oxford (by 8 Jan 2025): ox.ac.uk/admissions/g...


