By focusing on minimal pathways, we can enumerate or sample pathways efficiently in large models with new methods. ⚡
This also enables some cool new applications: 🧵
arxiv.org/abs/2511.05906

arxiv.org/abs/2511.05906
www.nytimes.com/interactive/...

www.nytimes.com/interactive/...
Topic: Synergistic coevolution in mono-specific and multi-species microbial consortia
Please RT or forward this information to interested candidates.
Deadline: 11.01.26
More info:
shorturl.at/f1TuF

Topic: Synergistic coevolution in mono-specific and multi-species microbial consortia
Please RT or forward this information to interested candidates.
Deadline: 11.01.26
More info:
shorturl.at/f1TuF
Sponsors: @amiposts.bsky.social, Pendulum, & Liv (@zymoresearch.bsky.social)
Registrants will receive free memberships to Applied Microbiology International.
Details below 👇

Sponsors: @amiposts.bsky.social, Pendulum, & Liv (@zymoresearch.bsky.social)
Registrants will receive free memberships to Applied Microbiology International.
Details below 👇
apply.interfolio.com/177622
Suggested deadline: 12/15/2025.
@columbiasysbio.bsky.social

apply.interfolio.com/177622
Suggested deadline: 12/15/2025.
@columbiasysbio.bsky.social
a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...




a 🧵 1/n
Drain: arxiv.org/abs/2511.04820
Strain: direct.mit.edu/qss/article/...
Oligopoly: direct.mit.edu/qss/article/...

Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...

Metalog: curated and harmonised contextual data for global metagenomics samples
now out in @narjournal.bsky.social
academic.oup.com/nar/advance-...



go.nature.com/4gzhLCo

go.nature.com/4gzhLCo
It converts Markdown and LaTeX to Unicode that can be used in “tweets”, and automatically splits long threads. Try it out!
keenancrane.github.io/LaTweet/

It converts Markdown and LaTeX to Unicode that can be used in “tweets”, and automatically splits long threads. Try it out!
keenancrane.github.io/LaTweet/
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

We pay to publish
Pay to read
Write for free
Edit for free
Proofread for free
And they won’t even give us a single login across journals 🤪
We pay to publish
Pay to read
Write for free
Edit for free
Proofread for free
And they won’t even give us a single login across journals 🤪

PhD position in Evolutionary Systems Biology at Stockholm University & SciLifeLab
🎓 Focus: evolution, development, computational & mathematical modeling, prediction
📍 Stockholm, Sweden
🕒 4 years, fully-funded
🔍 Details: su.varbi.com/en/what:job/...
Please share!

PhD position in Evolutionary Systems Biology at Stockholm University & SciLifeLab
🎓 Focus: evolution, development, computational & mathematical modeling, prediction
📍 Stockholm, Sweden
🕒 4 years, fully-funded
🔍 Details: su.varbi.com/en/what:job/...
Please share!
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
www.pnas.org/doi/abs/10.1...
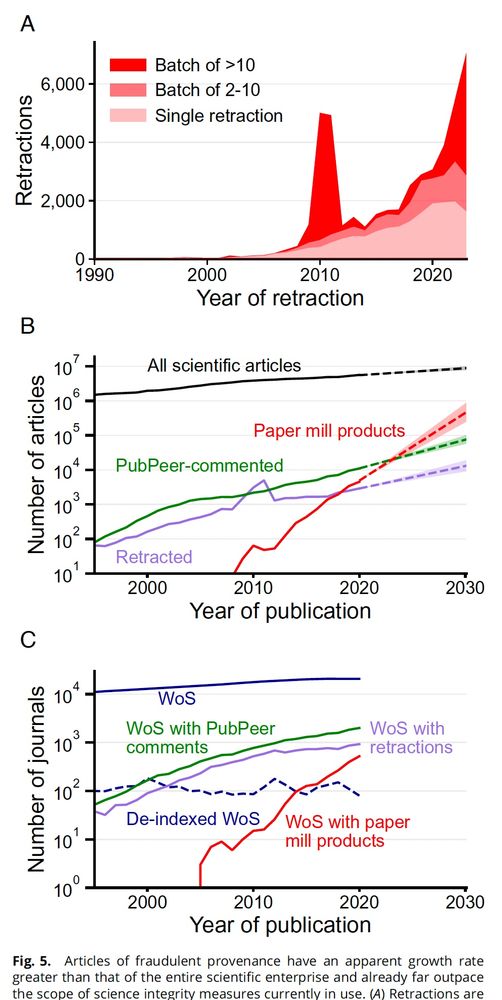
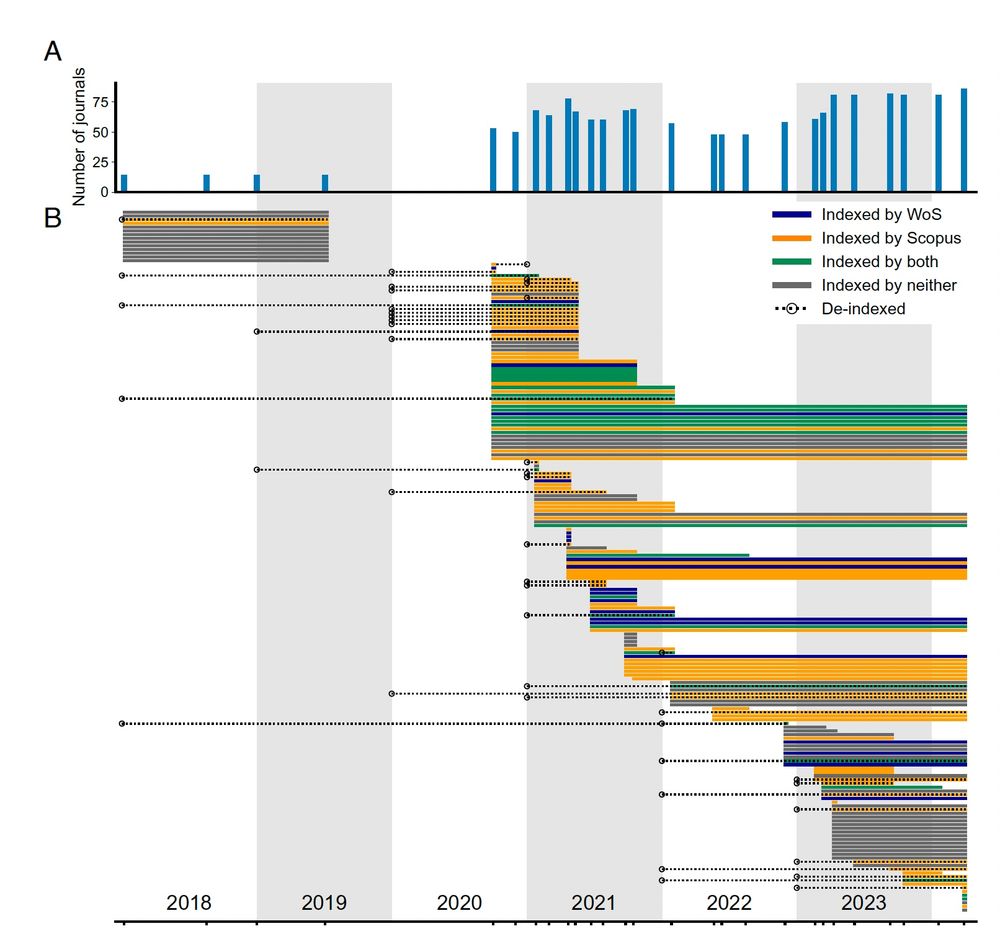
www.pnas.org/doi/abs/10.1...
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
Excellent work by UW Master's student Gechlang Tang in @asm.org #mSystems Journal.
journals.asm.org/doi/10.1128/...
🧵
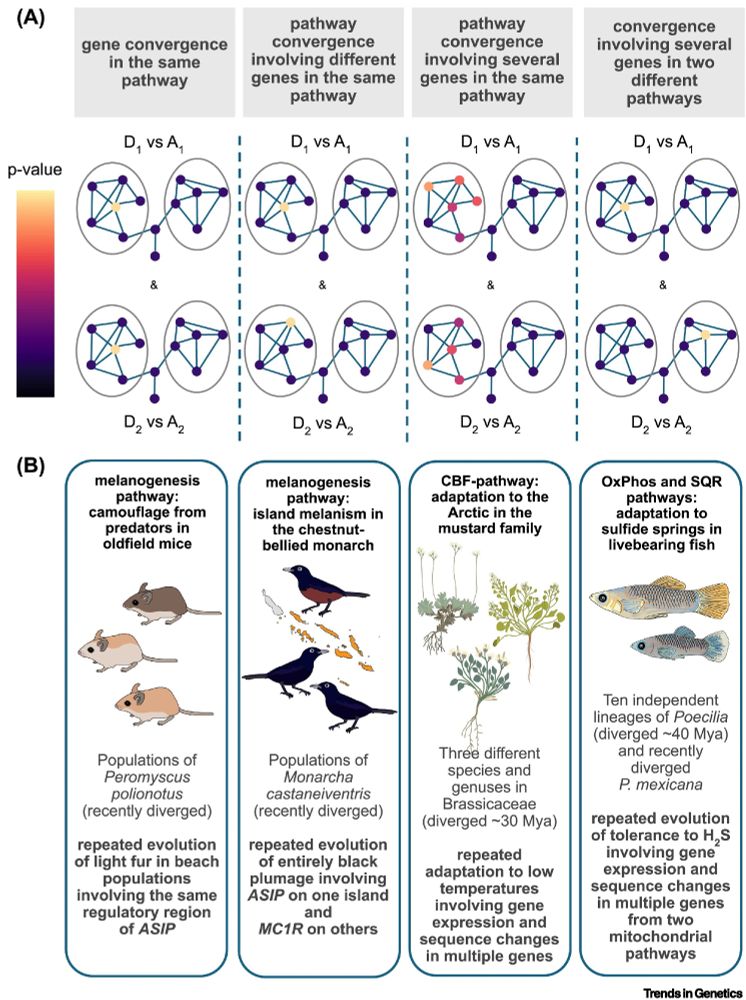
by Peter Hoitinga (@peter-hoitinga.bsky.social)
& Siri Birkeland (@siribirkeland.bsky.social)
"The extent to which evolution is repeatable at the genetic level remains a central question in biology..."
shorturl.at/NXF4k