
Collaboration, coding & discovery

Collaboration, coding & discovery

What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
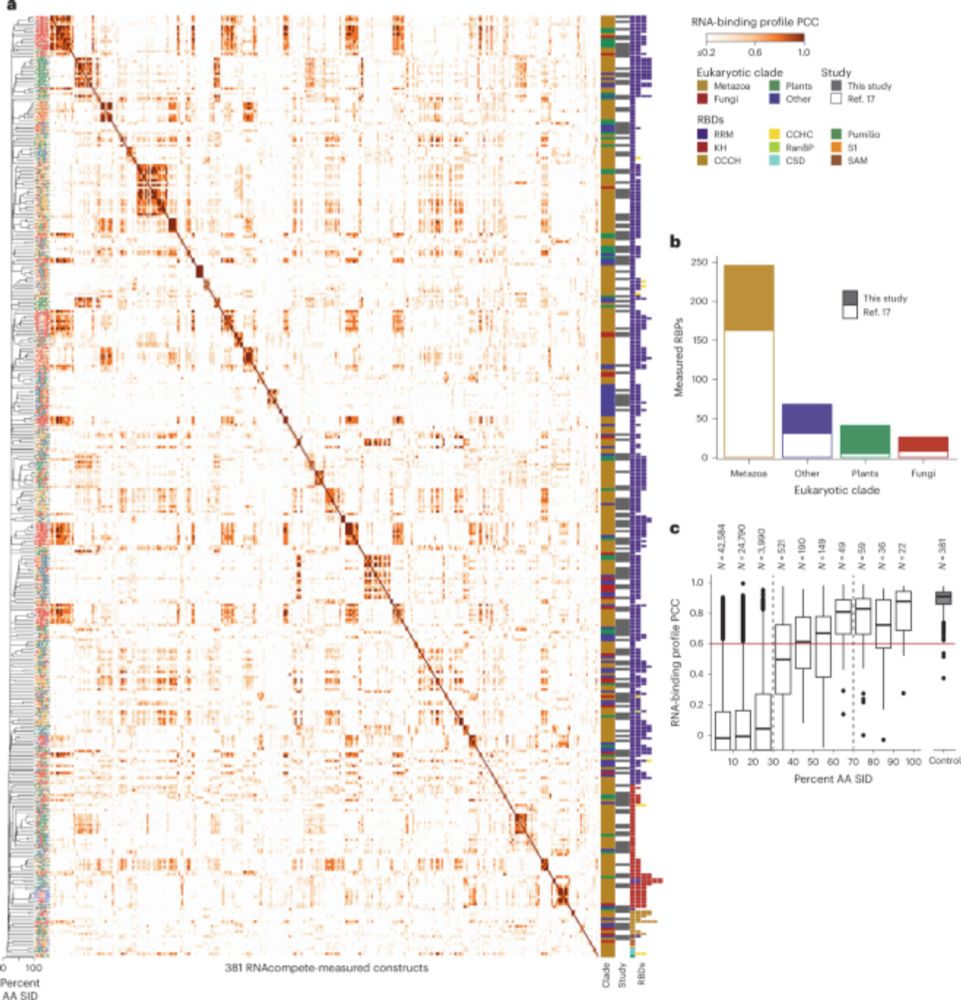
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
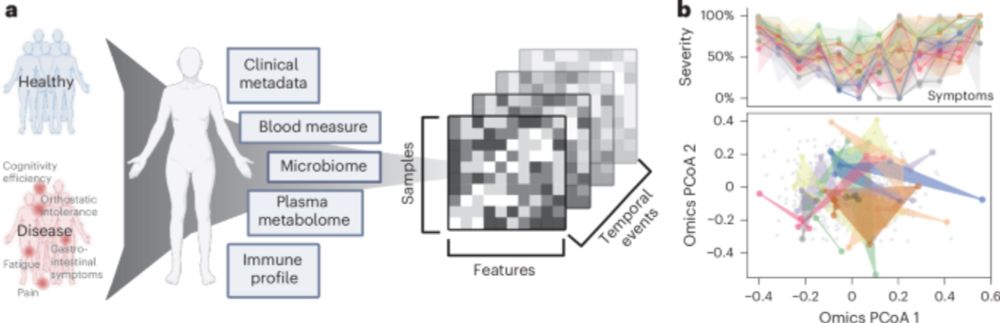
www.nature.com/articles/s41...
www.ft.com/content/9cdf...
www.nature.com/articles/s41...
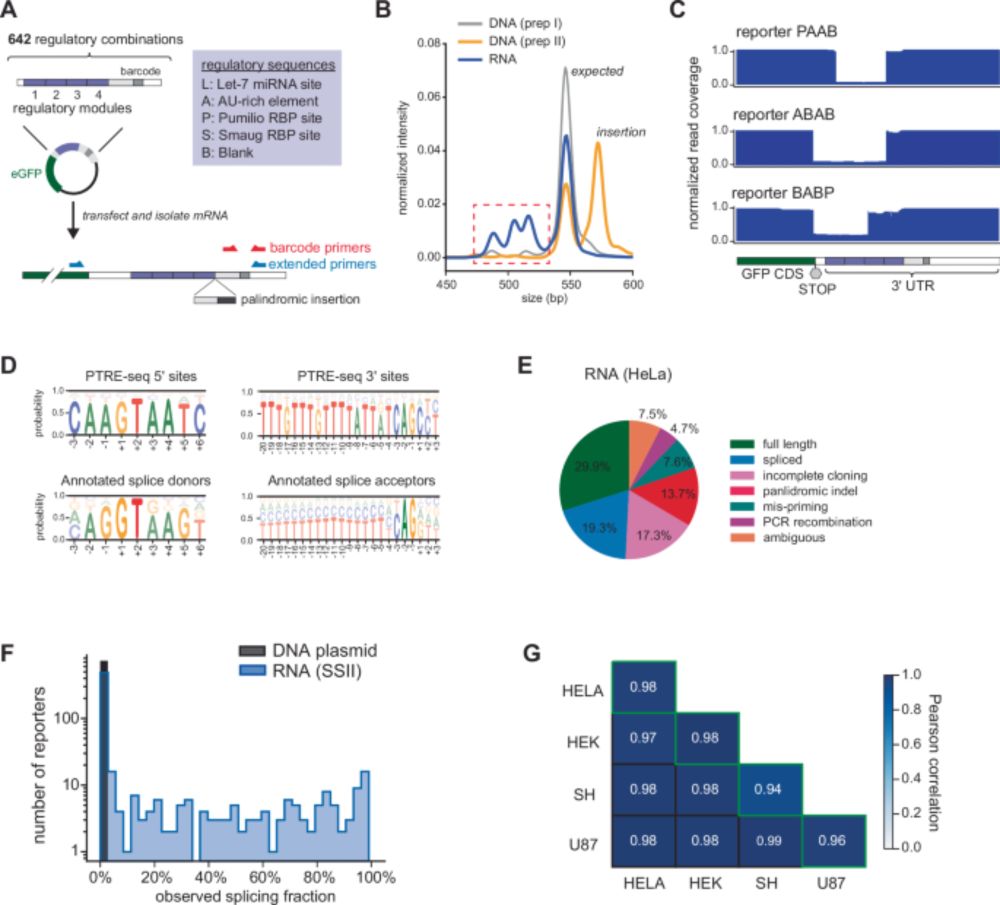
www.nature.com/articles/s41...
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
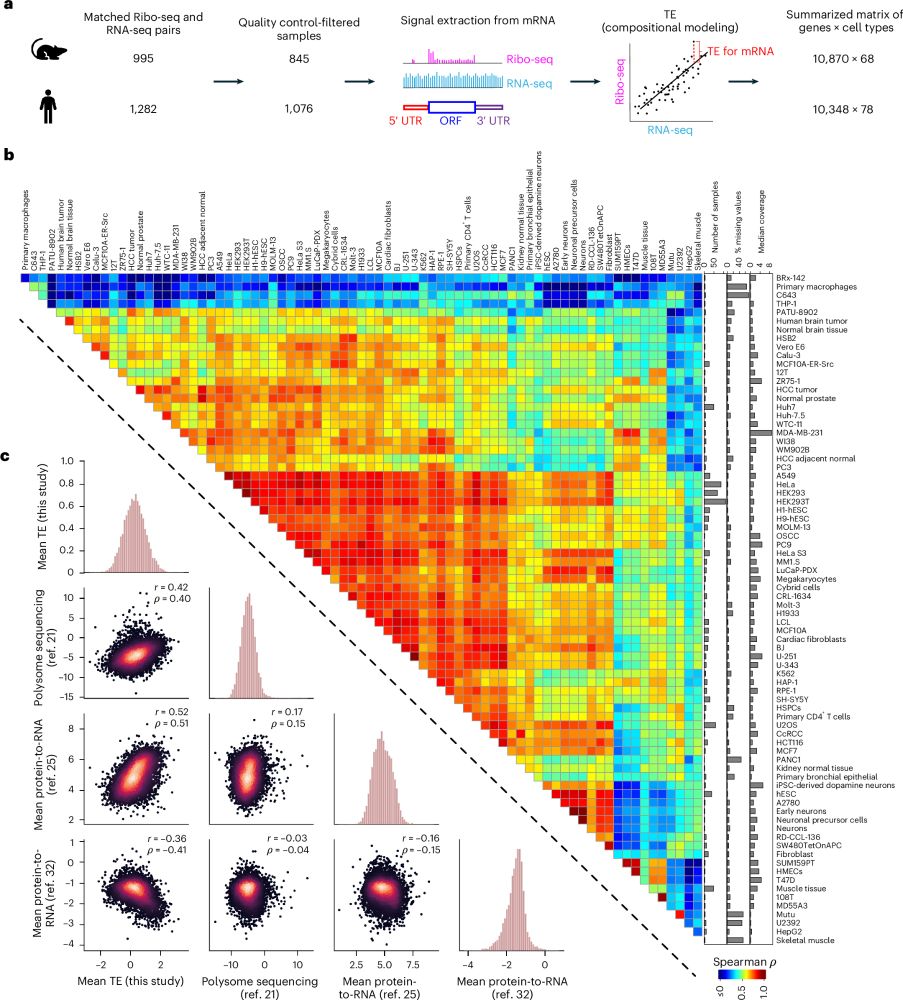
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org

Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org

Spoiler: regulation is key

Spoiler: regulation is key







#RegSys #ismbeccb2025 @ferhatay.bsky.social

#RegSys #ismbeccb2025 @ferhatay.bsky.social
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social

@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social


