Marcel Schulz
@themarcelschulz.bsky.social
120 followers
290 following
22 posts
Director of Institute for Computational Genomic Medicine at Goethe University Frankfurt https://cgm.uni-frankfurt.de/
Posts
Media
Videos
Starter Packs
Reposted by Marcel Schulz
Reposted by Marcel Schulz
Reposted by Marcel Schulz
Reposted by Marcel Schulz
Reposted by Marcel Schulz
Eric Topol
@erictopol.bsky.social
· Jul 25
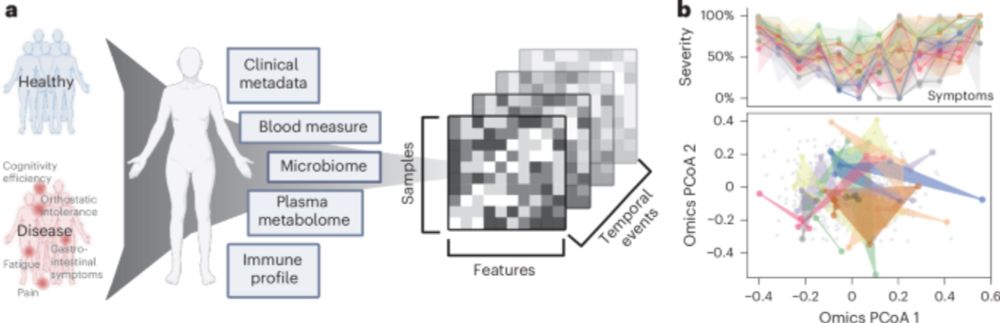
AI-driven multi-omics modeling of myalgic encephalomyelitis/chronic fatigue syndrome - Nature Medicine
Analysis of a deeply phenotyped, longitudinal cohort of 249 participants elucidates the network of concurrent molecular and macroscopic patterns related to insurgence and course of ME/CFS.
www.nature.com
Reposted by Marcel Schulz
Reposted by Marcel Schulz
Anthony Mathelier
@amathelier.bsky.social
· Jul 24
Reposted by Marcel Schulz
Anthony Mathelier
@amathelier.bsky.social
· Jul 24
Reposted by Marcel Schulz
Reposted by Marcel Schulz