
🔗 cd-code.org
🔗 cd-code.org
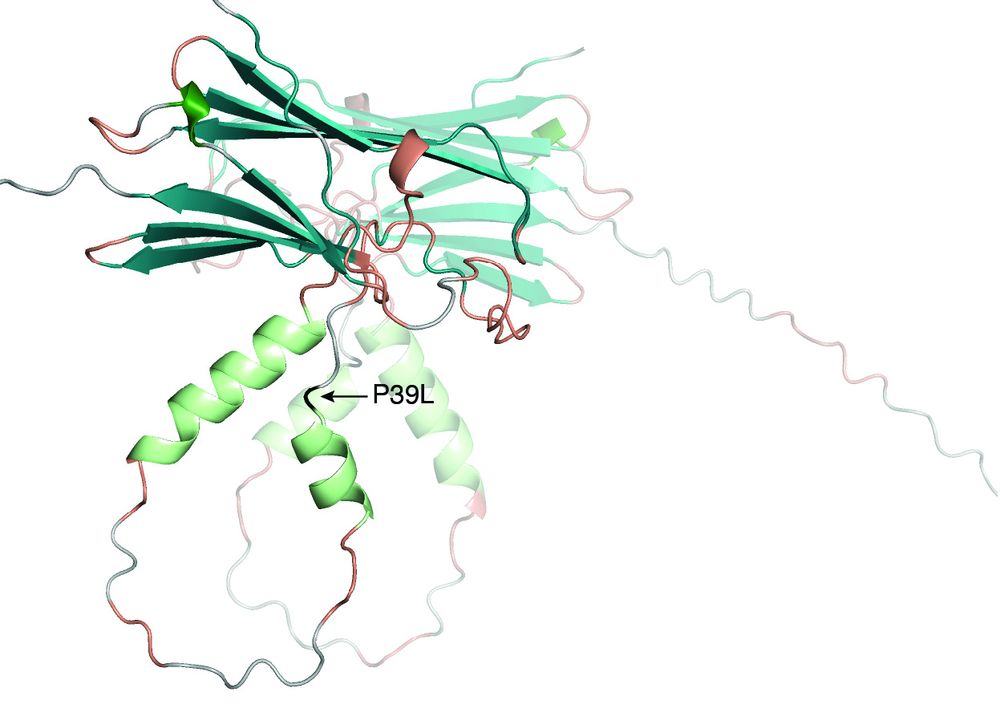
CD-CODE 2.0 stores #biomolecular #condensates and their #protein and nucleic acid components and now includes repository of 🦠 infectious condensates, 💊 condensate-modulating compounds, and 🩺 condensatopathies
👀 doi.org/10.1093/nar/...


Commentary in @natnano.nature.com
#nanotechnology

Commentary in @natnano.nature.com
#nanotechnology
▶️ @ssp2025.bsky.social

▶️ @ssp2025.bsky.social

https://go.nature.com/40wXMx6

https://go.nature.com/40wXMx6
ℹ️ tu-dresden.de/tu-dresden/n...

Big #thankyou to all the participants from
@hymanlab.bsky.social, @tothpetroczylab.bsky.social and von Appen labs for your support and enthusiasm!💜
Big #thankyou to all the participants from
@hymanlab.bsky.social, @tothpetroczylab.bsky.social and von Appen labs for your support and enthusiasm!💜
doi.org/10.1093/nar/...
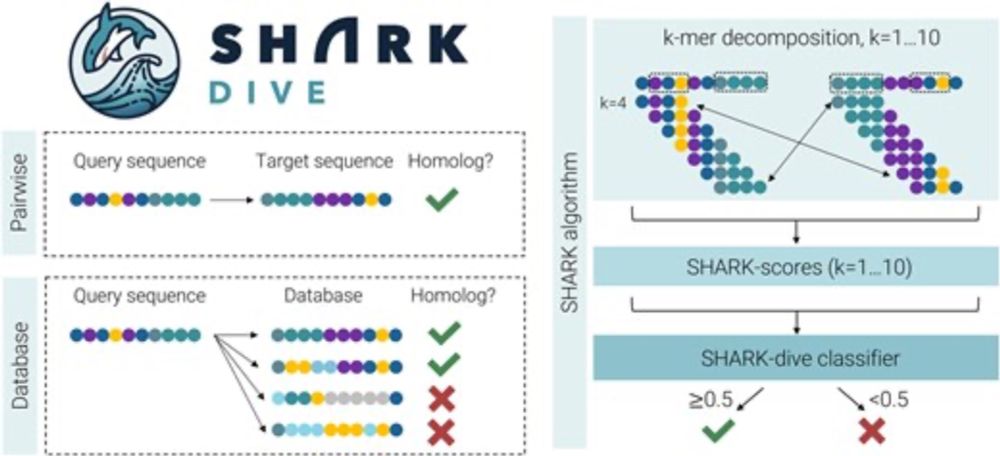
doi.org/10.1093/nar/...
Learn more and apply: physics-of-life.tu-dresden.de/career-educa...
#internationalstudents #MSc #EnglishMSc #Physics #studyabroad
✅ €300/month for 1 year
✅ No separate application—admitted students are automatically considered.
Apply now ➡️ tud.link/vejrmx
#Biophysics #PhysicsofLife #StudyinGermany #Masters #Biology

Learn more and apply: physics-of-life.tu-dresden.de/career-educa...
#internationalstudents #MSc #EnglishMSc #Physics #studyabroad
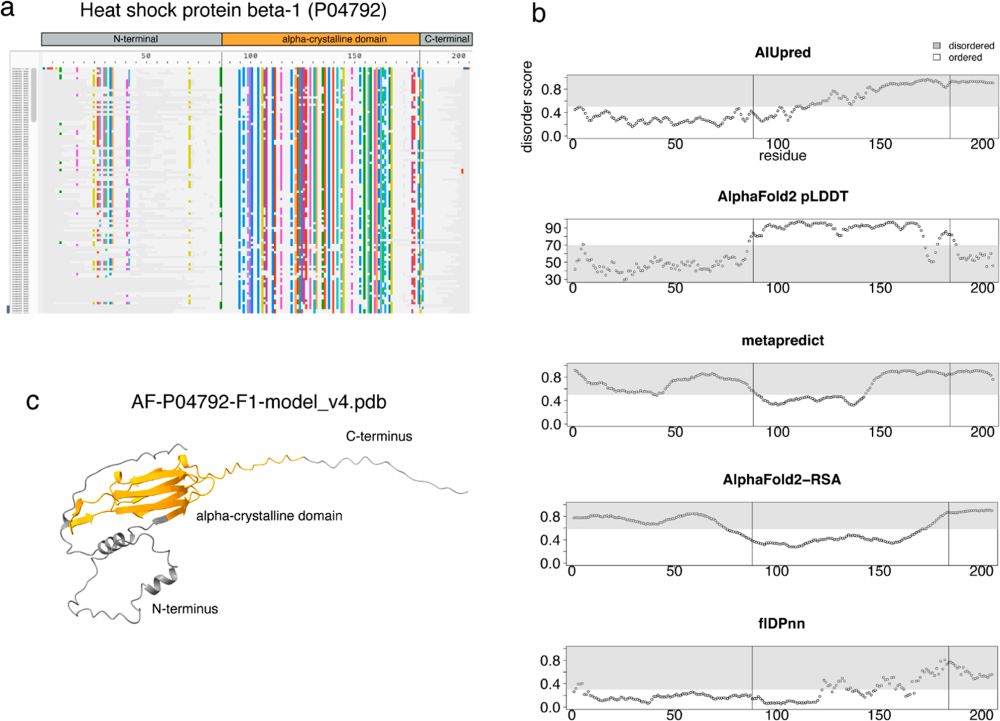
It's a great step forward, directing further developments and outlook for our project @tothpetroczylab.bsky.social @mscheremetjew.bsky.social 🥳
Spreading knowledge about #biomolecular #condensates 🥳
@ebi.embl.org @sib.swiss @mpi-cbg.de @tothpetroczylab.bsky.social @hymanlab.bsky.social

It's a great step forward, directing further developments and outlook for our project @tothpetroczylab.bsky.social @mscheremetjew.bsky.social 🥳

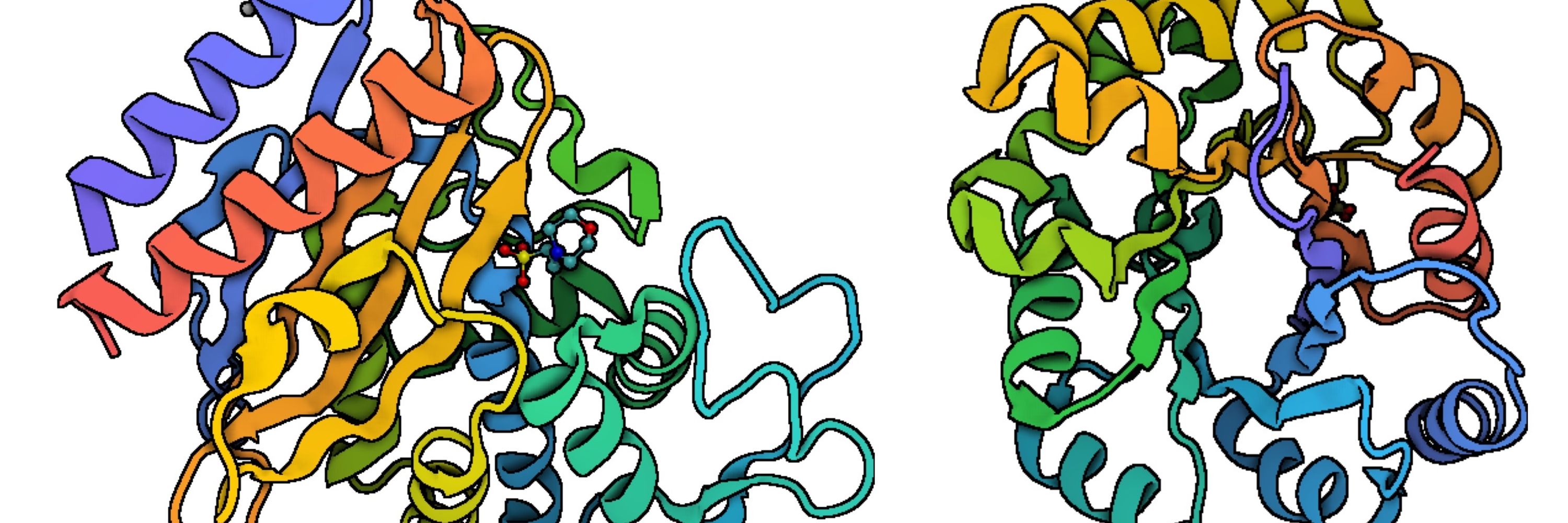
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart

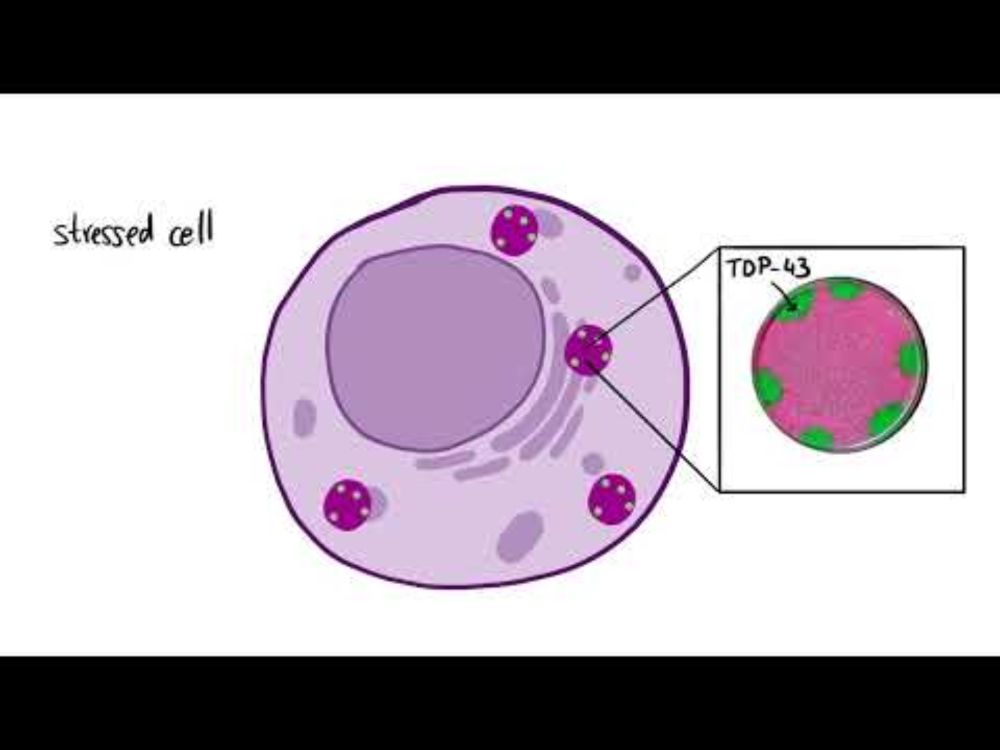
👀Check out the new #biomolecular #condensates at cd-code.org
👀We're continuing to update #Protein - #condensate connections and resolve issues!
Join us to share your #condensate #expertise as a contributor: cd-code.org/signup
#Bioinformatics #DatabaseUpdate #IDR

👀Check out the new #biomolecular #condensates at cd-code.org
👀We're continuing to update #Protein - #condensate connections and resolve issues!
Join us to share your #condensate #expertise as a contributor: cd-code.org/signup
#Bioinformatics #DatabaseUpdate #IDR



