journals.plos.org/plosbiology/...

journals.plos.org/plosbiology/...
@umich.edu #MedSky #IDSky

@umich.edu #MedSky #IDSky
This was *so much fun*. I was nervous going into it (I really look up to Steve!), but I had a blast and I think this is the best interview I have ever done.
Thanks for having me on, folks!
www.quantamagazine.org/how-did-mult...

This was *so much fun*. I was nervous going into it (I really look up to Steve!), but I had a blast and I think this is the best interview I have ever done.
Thanks for having me on, folks!
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
With mentorship from the amazing @ksxue.bsky.social, I looked at how the outcomes of species introductions to microbial communities are influenced by the number of introduced microbes.
Takehome: Widespread and substantial deceleration in fitness with age!
Amazing effort by PhD student Hamish MacGregor 💪
www.biorxiv.org/content/10.1...
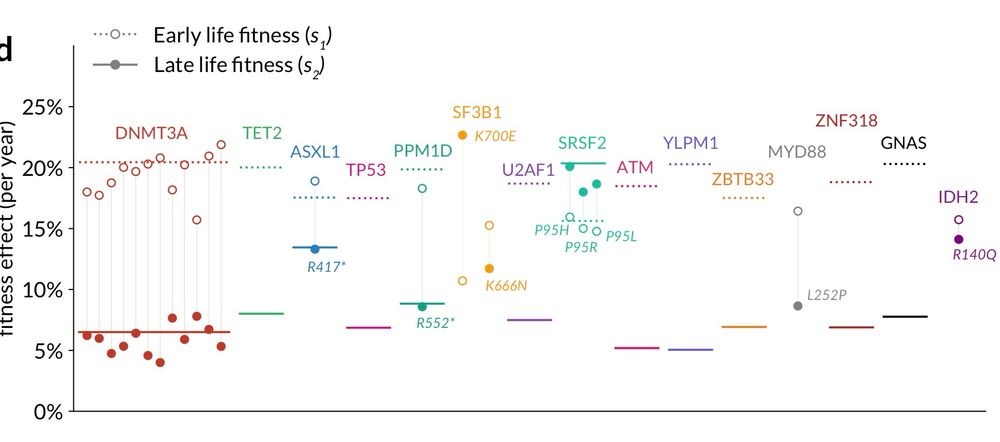
Takehome: Widespread and substantial deceleration in fitness with age!
Amazing effort by PhD student Hamish MacGregor 💪
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Learn more in our new paper 👉 rdcu.be/d07Np

Learn more in our new paper 👉 rdcu.be/d07Np
urldefense.com/v3/__https:/...

urldefense.com/v3/__https:/...
Here's a link to my notes on population & quantitative genetics:
github.com/cooplab/popg...
Hoping to extend it more after the winter holidays, as I'm just finishing up teaching the undergrad version of class.
Here's a link to my notes on population & quantitative genetics:
github.com/cooplab/popg...
Hoping to extend it more after the winter holidays, as I'm just finishing up teaching the undergrad version of class.

This approach is quite flexible, and we're hoping to extend it to more complex scenarios, including N(t) and certain forms of positive selection!

This approach is quite flexible, and we're hoping to extend it to more complex scenarios, including N(t) and certain forms of positive selection!

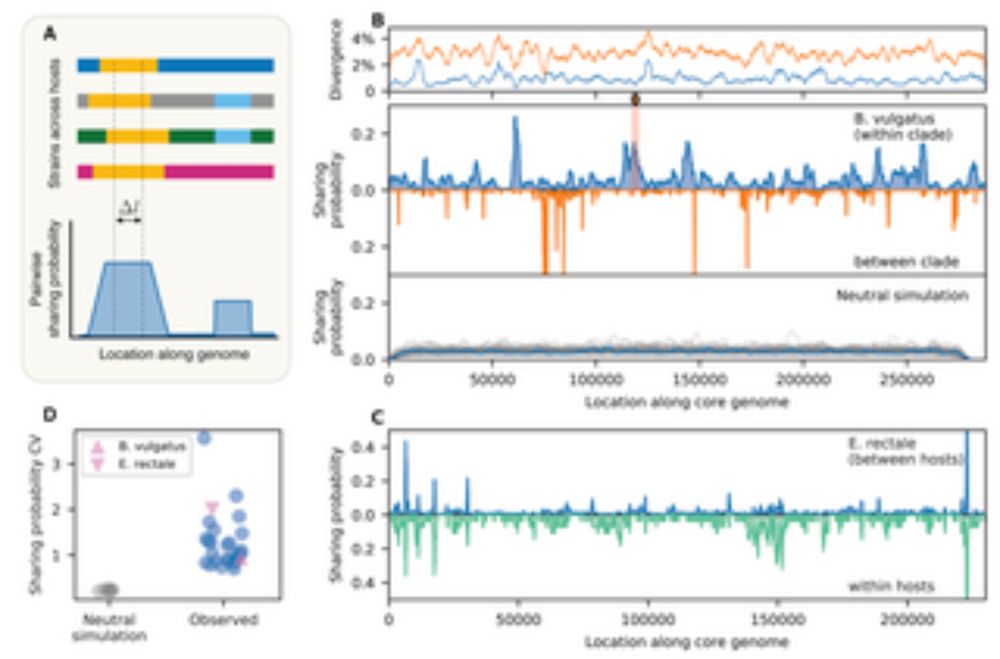
What do ecology and evolution look like in a 20-year microbiome time series? They blur together
@quendi.bsky.social @archaeal.bsky.social @uslter.bsky.social @sarilog.bsky.social 🧪🖥️🧬
www.biorxiv.org/content/10.1...

What do ecology and evolution look like in a 20-year microbiome time series? They blur together
@quendi.bsky.social @archaeal.bsky.social @uslter.bsky.social @sarilog.bsky.social 🧪🖥️🧬
www.biorxiv.org/content/10.1...
Anurag Limdi, Alex Couce, @relenski.bsky.social, and
Olivier Tenaillon exploring this over 50,000 generations of evolution is out today! 1/
(cross-post from the other place)
www.science.org/doi/10.1126/...

Anurag Limdi, Alex Couce, @relenski.bsky.social, and
Olivier Tenaillon exploring this over 50,000 generations of evolution is out today! 1/
(cross-post from the other place)
www.science.org/doi/10.1126/...
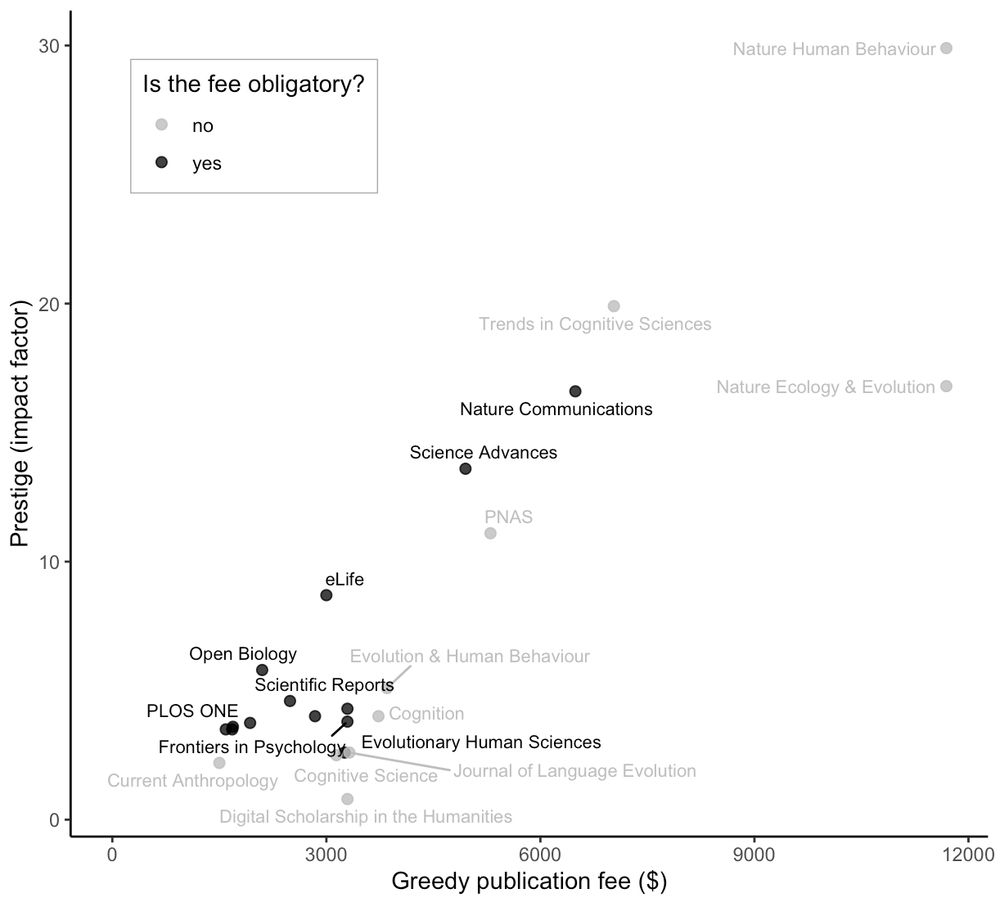
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Click Automated Services in the dashboard to view these. We welcome feedback. connect.biorxiv.org/news/2023/11...

Click Automated Services in the dashboard to view these. We welcome feedback. connect.biorxiv.org/news/2023/11...

Some deep sea hermit species live symbiotically with anemones that DISSOLVE the shell and SECRETE a new one ("carcinoecium") made of chitin! The carcinoecium has evolved at least 3 times! Photo credits in alt text. @megdaly.bsky.social



Some deep sea hermit species live symbiotically with anemones that DISSOLVE the shell and SECRETE a new one ("carcinoecium") made of chitin! The carcinoecium has evolved at least 3 times! Photo credits in alt text. @megdaly.bsky.social


