Alessandro Vannini
@alessandrovannini.bsky.social
970 followers
680 following
4 posts
Head of Structural Biology Research Centre of Human Technopole, Milan
Professor of Integrative Structural Biology.
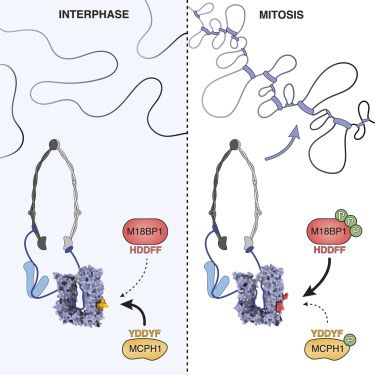
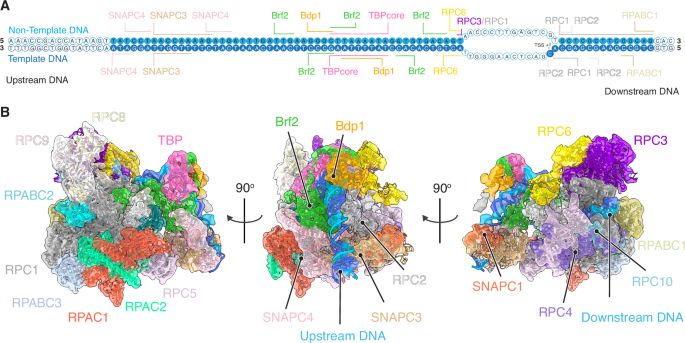
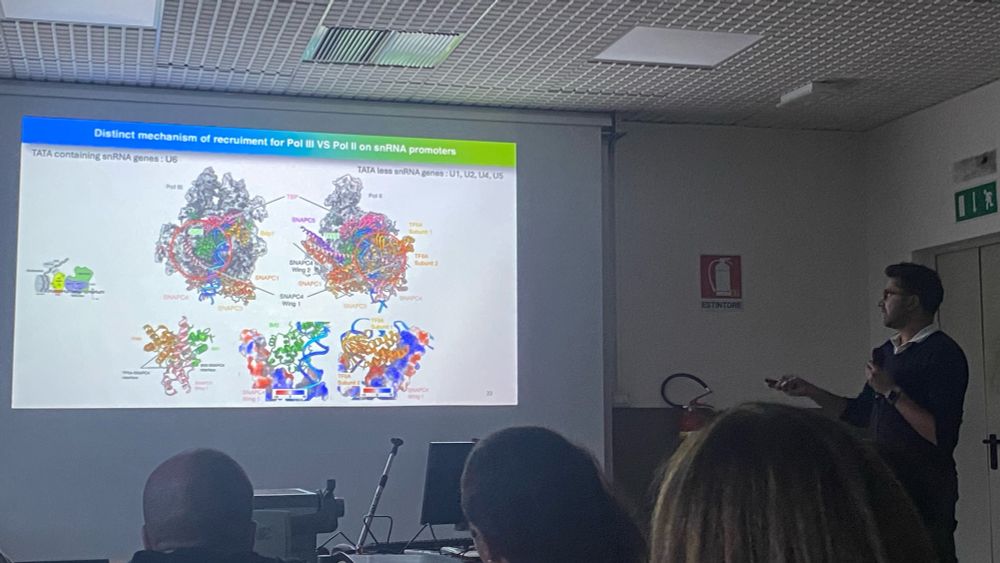
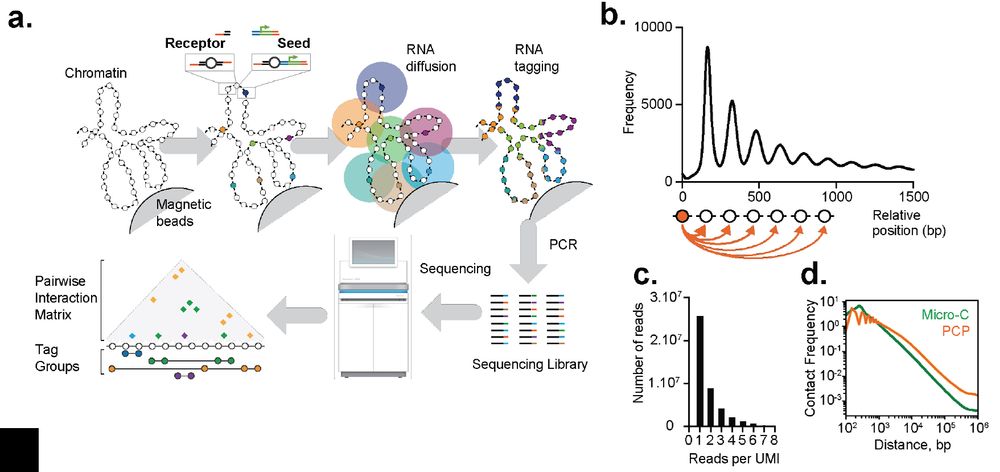
Our Team works on #RNAPolymeraseIII #SMC complexes and associated factors in genome function/organisation and viral sensing
Posts
Media
Videos
Starter Packs
Reposted by Alessandro Vannini
Reposted by Alessandro Vannini
Reposted by Alessandro Vannini
Reposted by Alessandro Vannini
Oliver Harschnitz
@harschnitz.bsky.social
· Nov 20
Reposted by Alessandro Vannini
Reposted by Alessandro Vannini
Reposted by Alessandro Vannini
Karim-Jean Armache
@kjarmache.bsky.social
· Nov 13
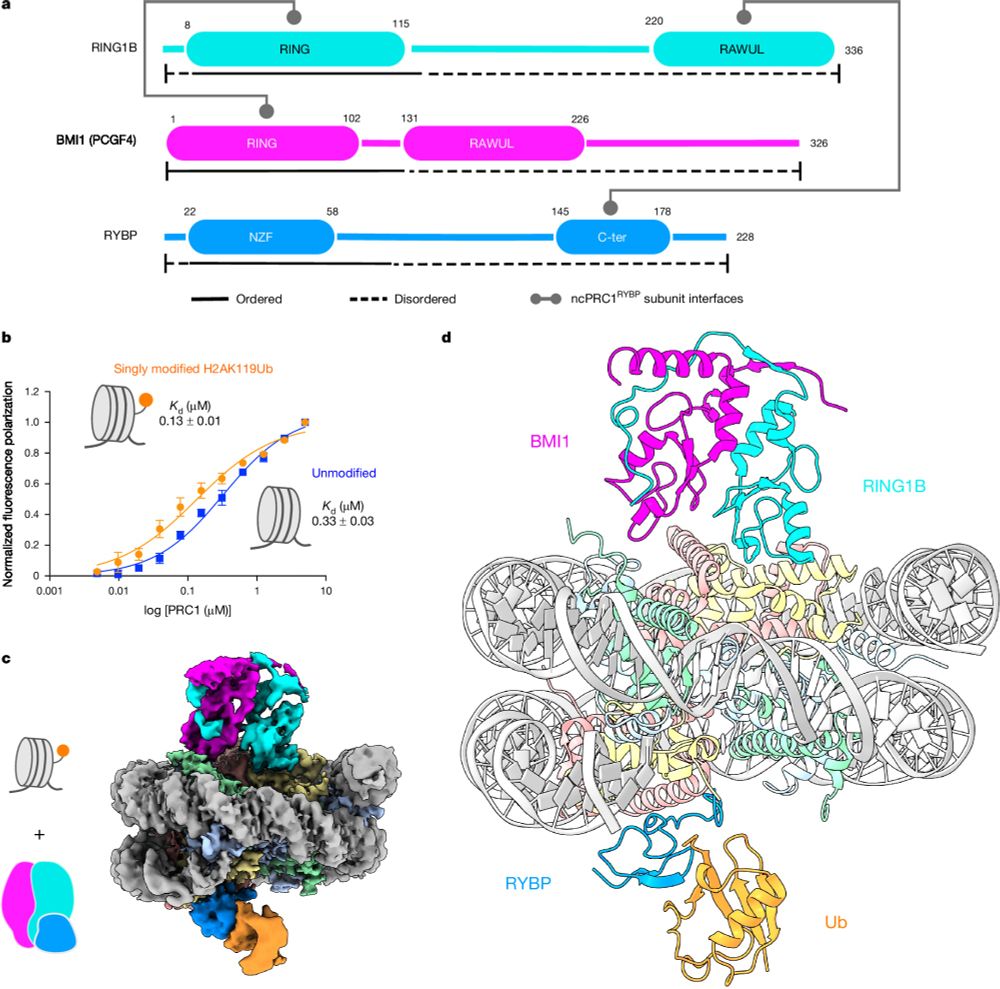
Read–write mechanisms of H2A ubiquitination by Polycomb repressive complex 1
Nature - Cryo-electron microscopy and biochemical studies elucidate the read–write mechanisms of non-canonical PRC1-containing RYBP in histone H2A lysine 119 monoubiquitination and their...
rdcu.be
Reposted by Alessandro Vannini
Rémi Fronzes
@fronzeslab.bsky.social
· Nov 13