Andrew Bowman
@bowchro.bsky.social
860 followers
750 following
76 posts
Scientist and serial hobbyist. Warwick Uni. How does chromatin move, and is it important? Contrary to my profile picture, I am ambivalent towards cats. Chickens however....
Posts
Media
Videos
Starter Packs
Reposted by Andrew Bowman
Andrew Bowman
@bowchro.bsky.social
· Aug 20
Andrew Bowman
@bowchro.bsky.social
· Aug 7

How thoughtful experimental design can empower biologists in the omics era - Nature Communications
Here, the authors discuss principles of experimental design that are relevant for all biology research, along with special considerations for projects using -omics approaches, highlighting common expe...
share.google
Reposted by Andrew Bowman
Reposted by Andrew Bowman
Reposted by Andrew Bowman
Andrew Bowman
@bowchro.bsky.social
· May 15
Andrew Bowman
@bowchro.bsky.social
· May 14
Reposted by Andrew Bowman
Reposted by Andrew Bowman
Rebecca Boyle 🌕
@rboyle31.bsky.social
· May 4
Andrew Bowman
@bowchro.bsky.social
· May 4
Andrew Bowman
@bowchro.bsky.social
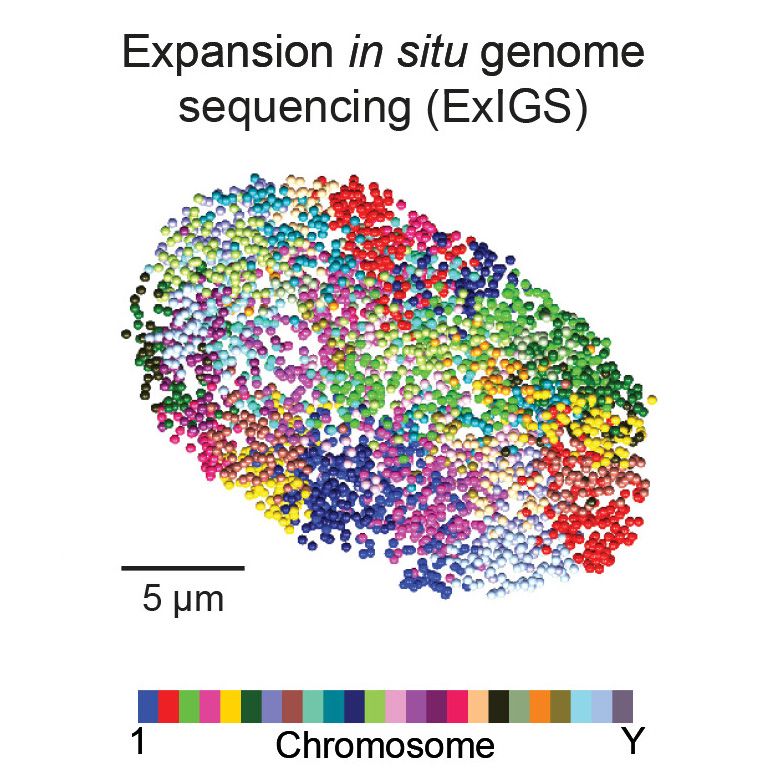
· Mar 28
Andrew Bowman
@bowchro.bsky.social
· Mar 10

In silico discovery of nanobody binders to a G-protein coupled receptor using AlphaFold-Multimer
Antibodies are central mediators of the adaptive immune response, and they are powerful research tools and therapeutics. Antibody discovery requires substantial experimental effort, such as immunization campaigns or in vitro library screening. Predicting antibody-antigen binding a priori remains challenging. However, recent machine learning methods raise the possibility of in silico antibody discovery, bypassing or reducing initial experimental bottlenecks. Here, we report a virtual screen using AlphaFold-Multimer (AF-M) that prospectively identified nanobody binders to MRGPRX2, a G protein-coupled receptor (GPCR) and therapeutic target for the treatment of pseudoallergic inflammation and itch. Using previously reported nanobody-GPCR structures, we identified a set of AF-M outputs that effectively discriminate between interacting and non-interacting nanobody-GPCR pairs. We used these outputs to perform a prospective in silico screen, identified nanobodies that bind MRGPRX2 with high affinity, and confirmed activity in signaling and functional cellular assays. Our results provide a proof of concept for fully computational antibody discovery pipelines that can circumvent laboratory experiments. ### Competing Interest Statement E.P.H., J.S.S., J.D.H., K.J.S., and A.C.K. are co-inventors on a patent application for MRGPRX2 antagonist nanobodies. D.S.M. is an advisor for Dyno Therapeutics, Octant, Jura Bio, Tectonic Therapeutic, and Genentech, and is a co-founder of Seismic Therapeutic. J.C.W. is a co-founder of MOMA therapeutics, a company in which he has a financial interest. A.C.K. is a co-founder and consultant for biotechnology companies Tectonic Therapeutic and Seismic Therapeutic, and for the Institute for Protein Innovation, a non-profit research institute. J.S.S. is an advisor / investigator for Biogen. The remaining authors declare no competing interests.
www.biorxiv.org
Reposted by Andrew Bowman