Boyan Bonev
@boyanbonev.bsky.social
370 followers
200 following
24 posts
Group Leader at Helmholtz Center Munich * Brain Epigenomics
Posts
Media
Videos
Starter Packs
Pinned
Boyan Bonev
@boyanbonev.bsky.social
· Apr 4

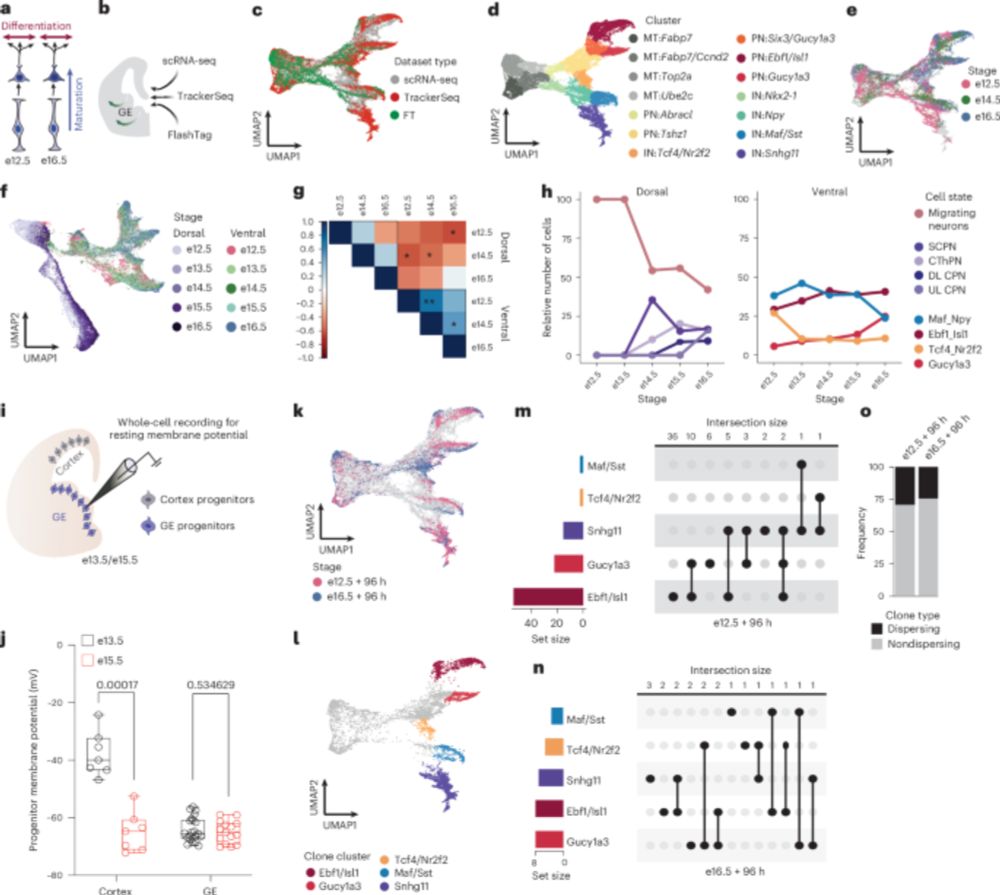
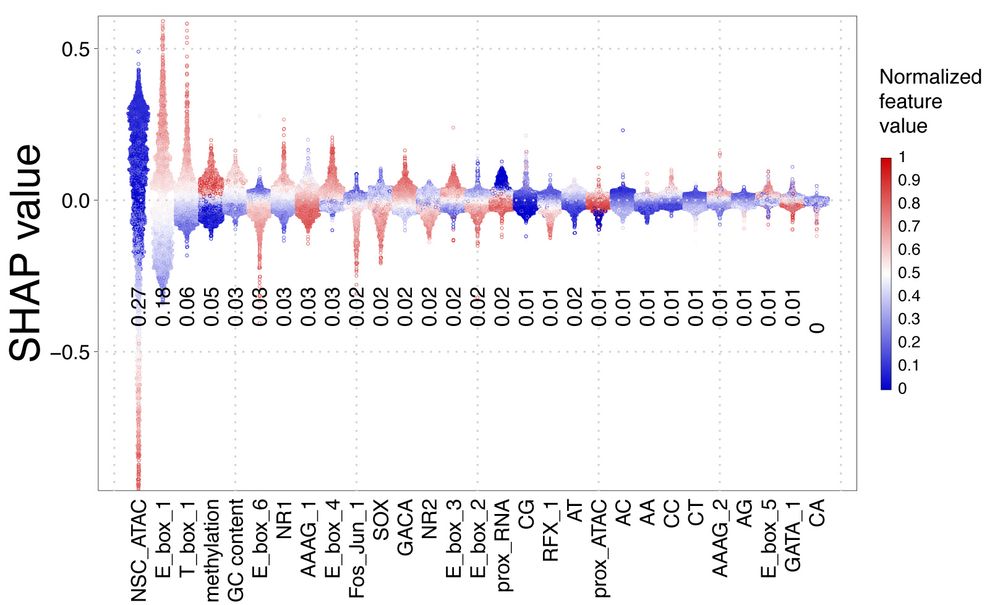
Neural stem cell epigenomes and fate bias are temporally coordinated during corticogenesis
The cerebral cortex orchestrates complex cognitive functions, yet how its distinct temporal lineages are molecularly patterned during development remains unresolved. Here, we integrate single-cell tra...
www.biorxiv.org
Reposted by Boyan Bonev
Hamperl Lab
@hamperllab.bsky.social
· Aug 4

Replisome progression regulates R-loop mediated transcriptional repression
Maintaining cellular proliferation necessitates the synchronized activity of diverse molecular machineries operating in parallel on the genome. Wide-spread transcription and R-loop formation can inter...
www.biorxiv.org
Reposted by Boyan Bonev
Reposted by Boyan Bonev
Boyan Bonev
@boyanbonev.bsky.social
· May 15
Reposted by Boyan Bonev
Lars Velten
@larsplus.bsky.social
· May 8

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
www.cell.com
Reposted by Boyan Bonev
Reposted by Boyan Bonev
Boyan Bonev
@boyanbonev.bsky.social
· Apr 4
Boyan Bonev
@boyanbonev.bsky.social
· Apr 4
Boyan Bonev
@boyanbonev.bsky.social
· Apr 4

Neural stem cell epigenomes and fate bias are temporally coordinated during corticogenesis
The cerebral cortex orchestrates complex cognitive functions, yet how its distinct temporal lineages are molecularly patterned during development remains unresolved. Here, we integrate single-cell tra...
www.biorxiv.org
Reposted by Boyan Bonev
Reposted by Boyan Bonev
Reposted by Boyan Bonev