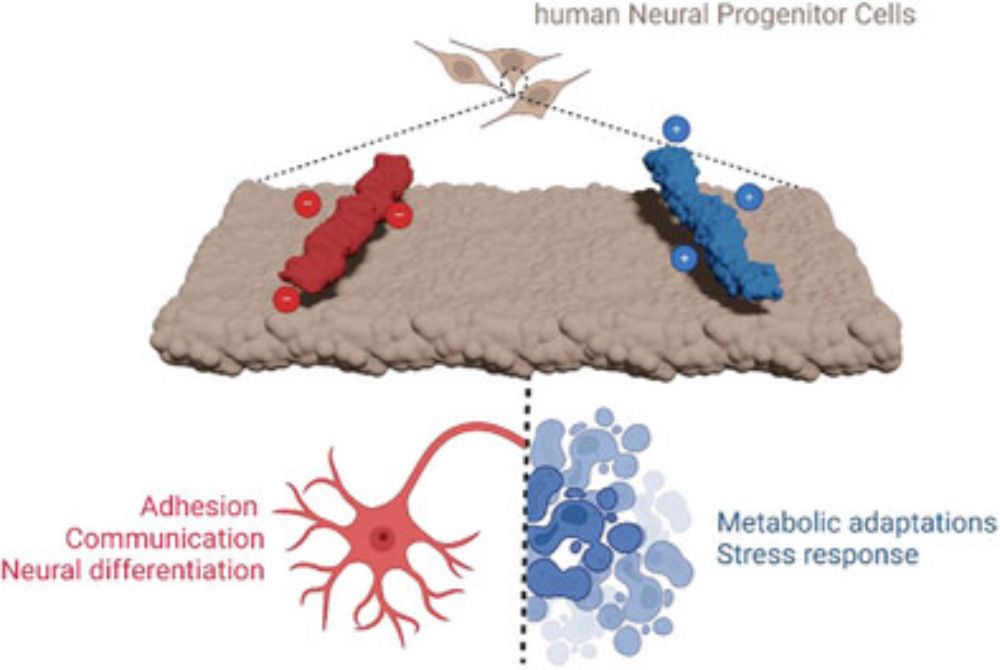
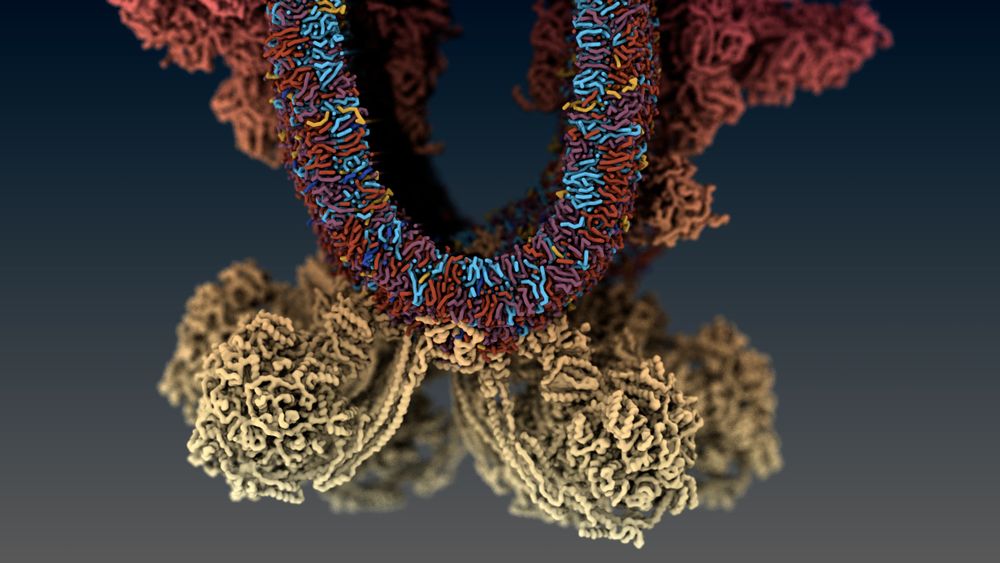
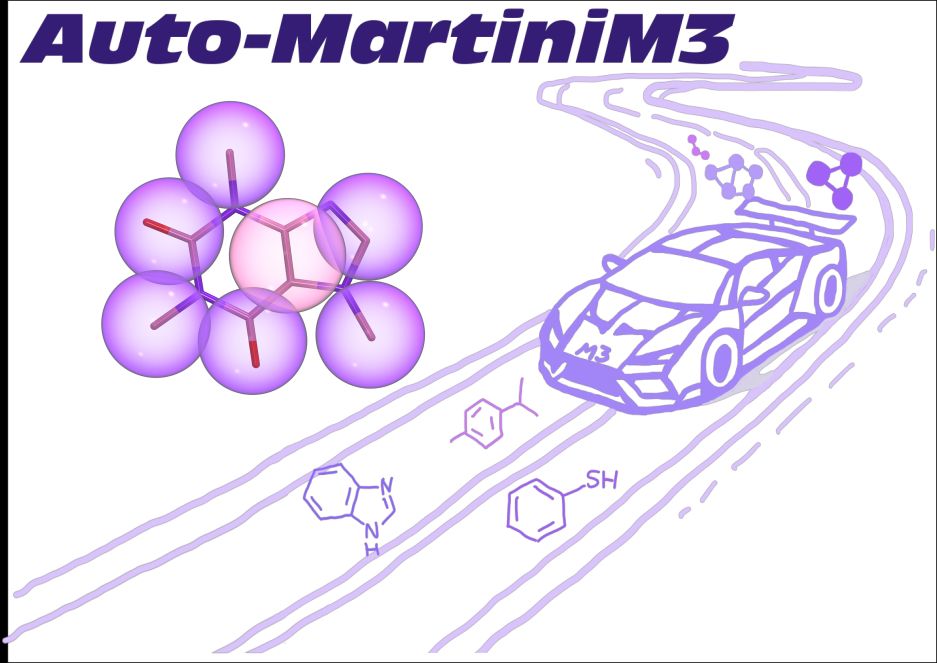
Siewert-Jan Marrink
@cg-martini.bsky.social
230 followers
0 following
31 posts
Martini lover.
Shaken, stirred, or self-assembled.
https://cgmartini.nl/
Posts
Media
Videos
Starter Packs
Reposted by Siewert-Jan Marrink
Reposted by Siewert-Jan Marrink
Reposted by Siewert-Jan Marrink
Reposted by Siewert-Jan Marrink
Reposted by Siewert-Jan Marrink
Reposted by Siewert-Jan Marrink