Dan Grabarczyk
@dangrabarczyk.bsky.social
490 followers
860 following
6 posts
Structural biologist - currently ubiquitin ligases, previously DNA replication and bacterial metabolism
Research Associate - Clausen Lab, IMP Vienna
Posts
Media
Videos
Starter Packs
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Fletcher Lab
@fletcherlab.bsky.social
· Sep 8

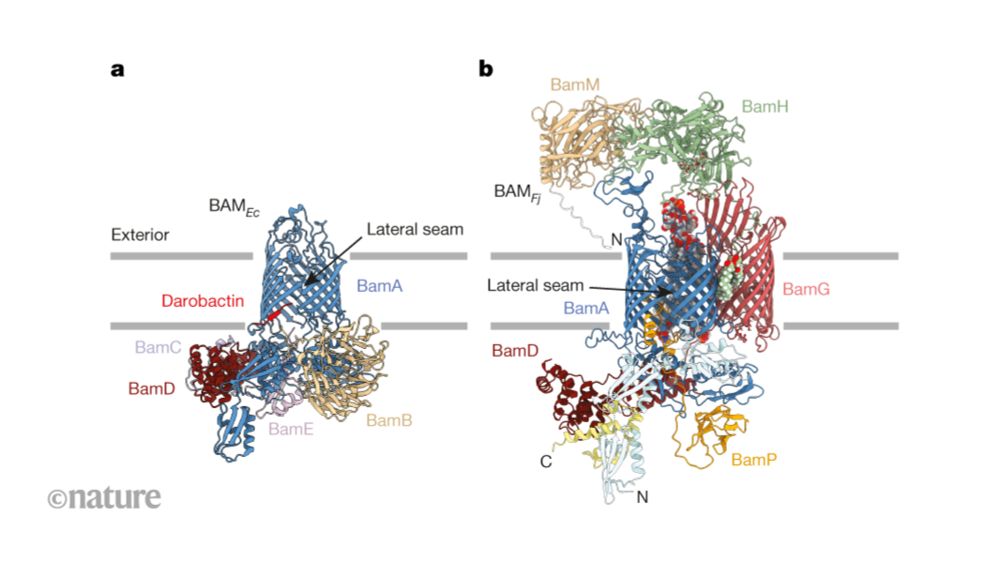
ZNFX1 uses two-component ubiquitin circuitry to quarantine viral RNA
The detection of viral RNA inside cells triggers a diverse range of antiviral responses, including global translation inhibition, interferon secretion and RNA sequestration. Mutations in the gene ZNFX...
www.biorxiv.org
Reposted by Dan Grabarczyk
IMP
@impvienna.bsky.social
· Aug 28

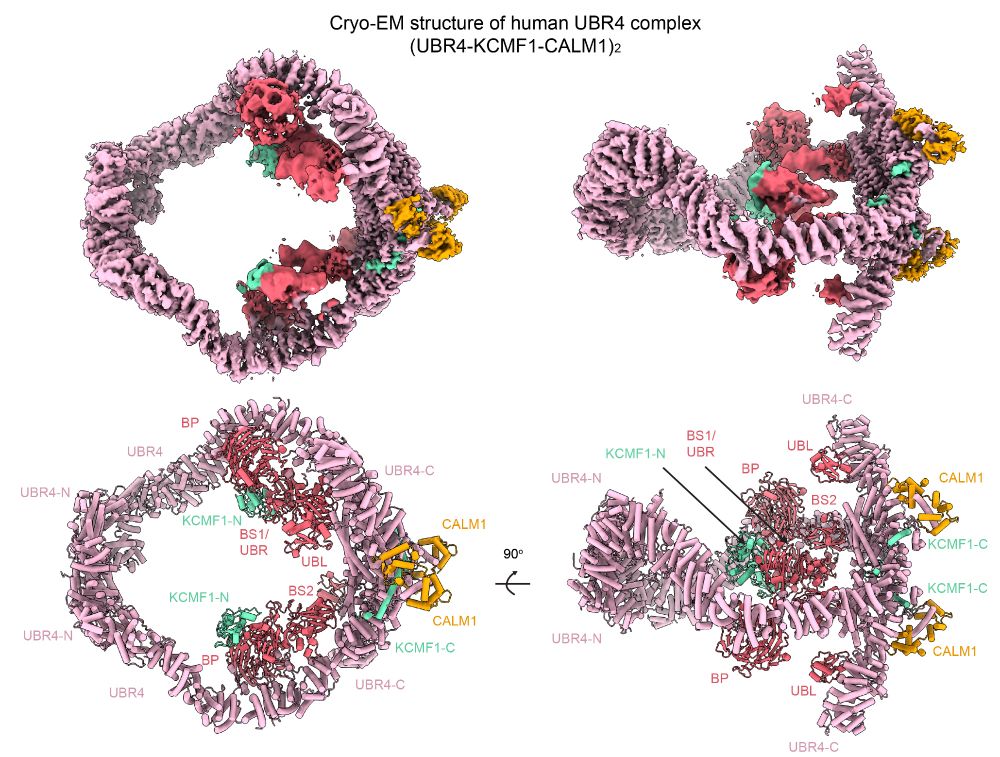
Inside the cell’s recycling hub: unveiling the architecture of UBR4
Researchers from the lab of Tim Clausen and collaborators have unveiled the three-dimensional structure of UBR4, a giant protein complex that safeguards cells by targeting defective proteins for destr...
www.imp.ac.at
Dan Grabarczyk
@dangrabarczyk.bsky.social
· Aug 28
Reposted by Dan Grabarczyk
IMP
@impvienna.bsky.social
· Aug 28

ZNFX1 compacts and tags RNA to keep immunity in check
Tim Clausen’s lab at the IMP and collaborators have uncovered how the ancient immune protein ZNFX1 enables cells to walk the fine line between fighting infection and avoiding autoimmune damage. The te...
www.imp.ac.at
Dan Grabarczyk
@dangrabarczyk.bsky.social
· Aug 27

A split-site E3 ligase mechanism enables ZNFX1 to ubiquitinate and cluster single-stranded RNA into ubiquitin-coated nucleoprotein particles
Grabarczyk et al. show the structure and mechanism of a non-canonical ubiquitin ligase,
which is activated through nucleic-acid-induced oligomerization and is critical for
cell survival during immune ...
www.cell.com
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
David B. Sauer
@davidbsauer.bsky.social
· Aug 15

Covalently constrained ‘Di-Gembodies’ enable parallel structure solutions by cryo-EM - Nature Chemical Biology
Disulfide-based dimerization of modified identical and heterologous nanobody scaffolds enables higher-order assembly for high-resolution cryo-electron microscopy structure determination that is widely...
www.nature.com
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Ewan Birney
@ewanbirney.bsky.social
· May 22

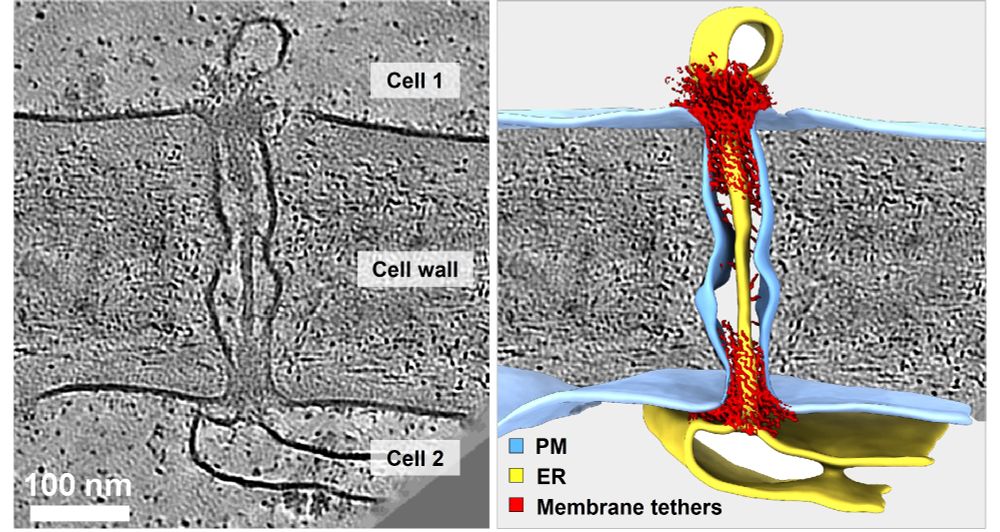
In-cell discovery and characterization of a non-canonical bacterial protein translocation-folding complex
Cryo-electron tomography has emerged as powerful technology for in-cell structural biology, and in combination with breakthroughs in protein structure prediction, offers a unique opportunity for illum...
www.biorxiv.org
Reposted by Dan Grabarczyk
Fletcher Lab
@fletcherlab.bsky.social
· May 13
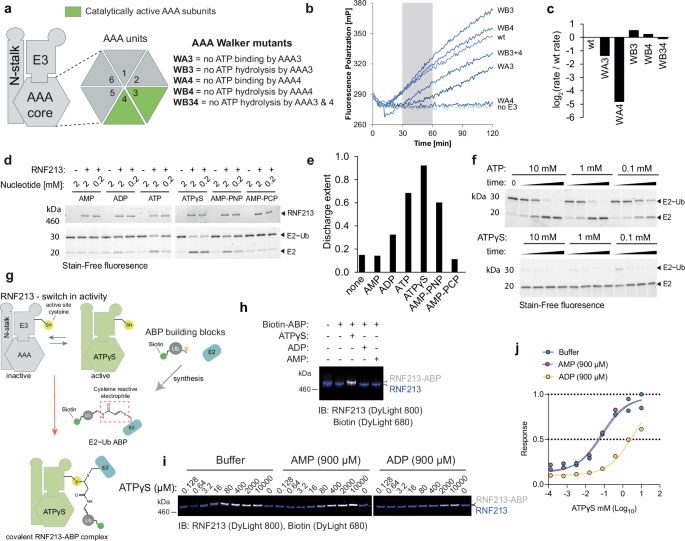
ATP functions as a pathogen-associated molecular pattern to activate the E3 ubiquitin ligase RNF213 - Nature Communications
RNF213 is an E3 ligase with ATPase activity. Here, the authors show that RNF213 is activated by ATP binding and senses cellular energy states, and reveal a transthiolation mechanism induced by immune ...
www.nature.com
Dan Grabarczyk
@dangrabarczyk.bsky.social
· May 10
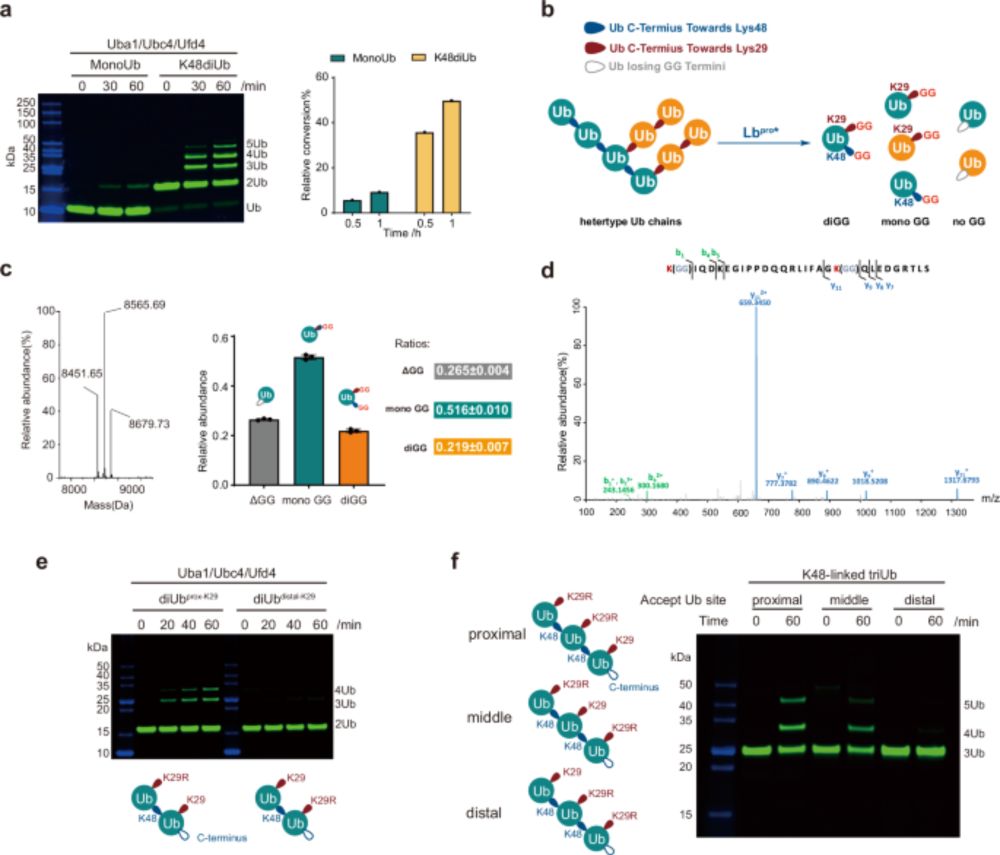
Structural visualization of HECT-type E3 ligase Ufd4 accepting and transferring ubiquitin to form K29/K48-branched polyubiquitination - Nature Communications
The K29/K48 hetero-polyubiquitin signal has been reported to be an enhanced degradation signal, but the enzymatic mechanism of this ubiquitin chain formation is unclear. Here, the authors reveal how t...
www.nature.com
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk
Reposted by Dan Grabarczyk