Dan Portik
@dportik.bsky.social
1.9K followers
720 following
79 posts
Bioinformatics scientist at PacBio. I currently work on metagenomics and methylation, but previously studied phylogenomics and frog/lizard evolution.
🦠 🐸 🦎 🧬
Free time: climbing 🧗, curling 🥌, and plants 🪴🌵.
Posts
Media
Videos
Starter Packs
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Reposted by Dan Portik
Dan Portik
@dportik.bsky.social
· Jun 11
Segata Lab
@cibiocm.bsky.social
· Jun 11

MetaPhlAn 4.2.2 release (initial long-read sequencing support and database update)
Announcement We are pleased to share that MetaPhlAn 4.2.2 is now available, which incorporates taxonomic profiling of long-read metagenomes for the first time and includes a new version of the MetaPhl...
forum.biobakery.org
Reposted by Dan Portik
Reposted by Dan Portik
Dan Portik
@dportik.bsky.social
· May 7
Dan Portik
@dportik.bsky.social
· Apr 22
Jon Belyeu
@jonbelyeu.bsky.social
· Apr 22

Complex structural variant visualization with SVTopo
Structural variants are genomic variants that impact at least 50 nucleotides and can play major roles in diversity and human health. Many structural variants are complex multi-breakpoint rearrangement...
www.biorxiv.org
Dan Portik
@dportik.bsky.social
· Apr 10
Chris Saunders
@ctsa.bsky.social
· Apr 10

Sawfish: Improving long-read structural variant discovery and genotyping with local haplotype modeling
AbstractMotivation. Structural variants (SVs) play an important role in evolutionary and functional genomics but are challenging to characterize. High-accu
doi.org
Reposted by Dan Portik
Reposted by Dan Portik
Guillaume Méric
@gmeric.bsky.social
· Feb 25
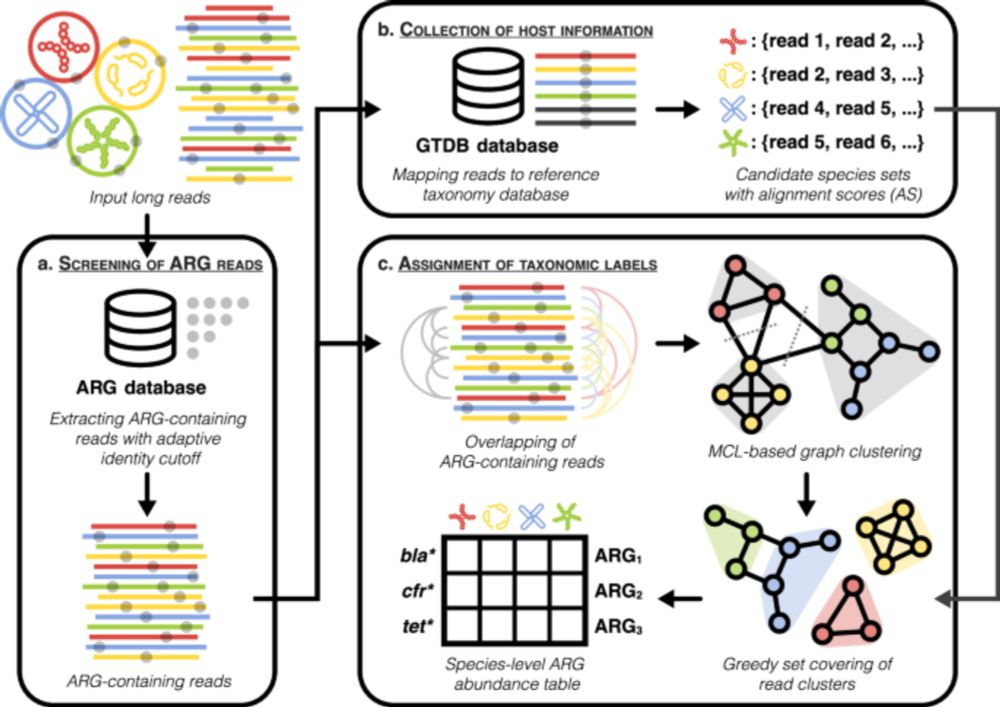
Species-resolved profiling of antibiotic resistance genes in complex metagenomes through long-read overlapping with Argo - Nature Communications
In this study, the authors present ‘Argo’, a bioinformatics tool for identifying the host species for antibiotic resistance genes in long-read metagenomic sequence data. They validate the tool using s...
www.nature.com
Dan Portik
@dportik.bsky.social
· Jan 31













